Nickel »
PDB 1a5n-1fwb »
1e9y »
Nickel in PDB 1e9y: Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid
Enzymatic activity of Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid
All present enzymatic activity of Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid:
3.5.1.5;
3.5.1.5;
Protein crystallography data
The structure of Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid, PDB code: 1e9y
was solved by
N.-C.Ha,
S.-T.Oh,
B.-H.Oh,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20 / 3.00 |
| Space group | I 2 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 178.160, 178.160, 178.160, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.3 / 28.2 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid
(pdb code 1e9y). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid, PDB code: 1e9y:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid, PDB code: 1e9y:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 1e9y
Go back to
Nickel binding site 1 out
of 2 in the Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid
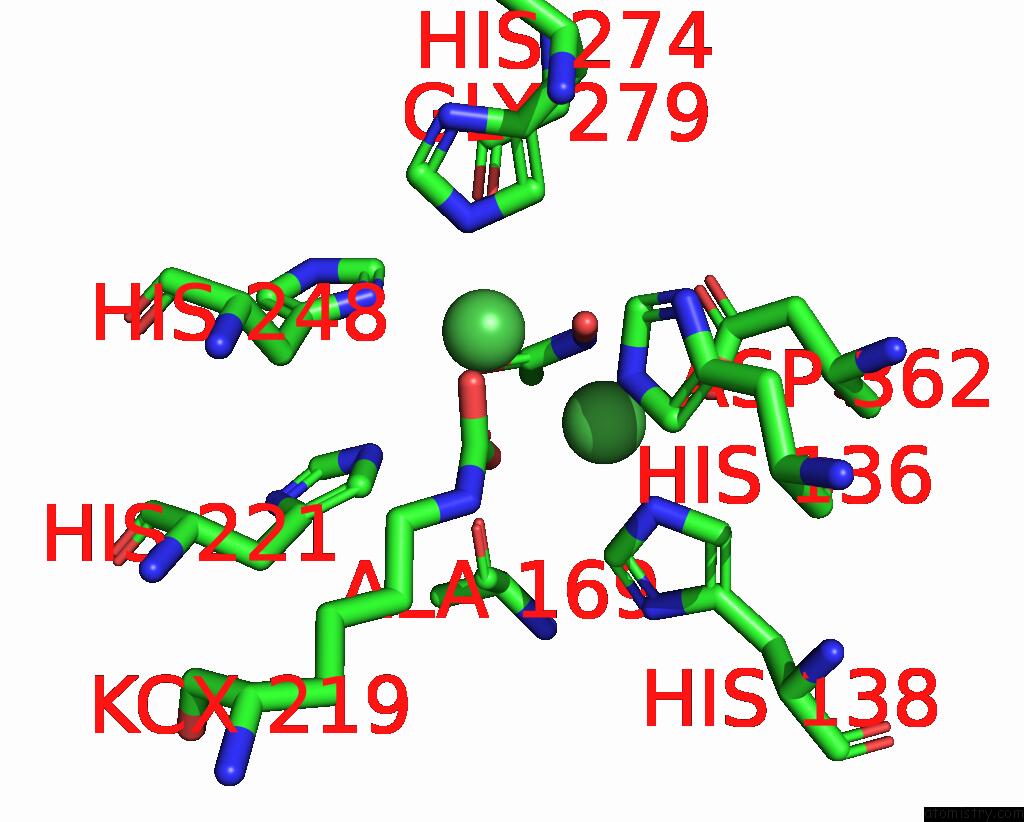
Mono view
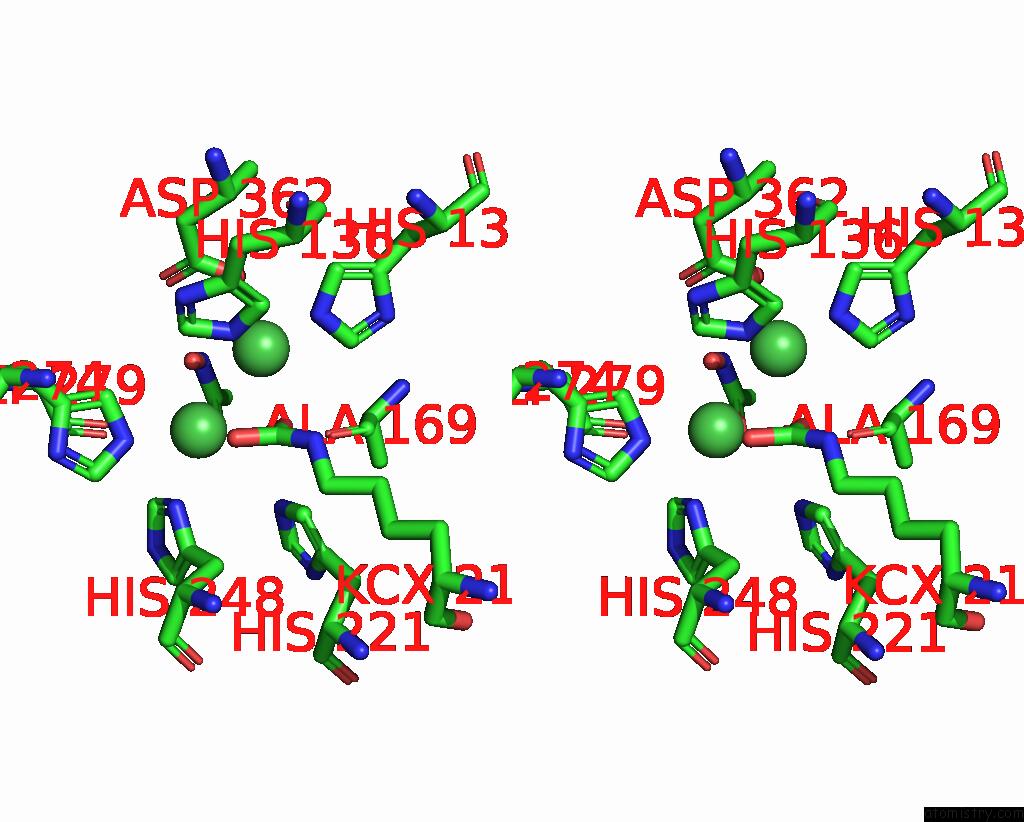
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid within 5.0Å range:
|
Nickel binding site 2 out of 2 in 1e9y
Go back to
Nickel binding site 2 out
of 2 in the Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid
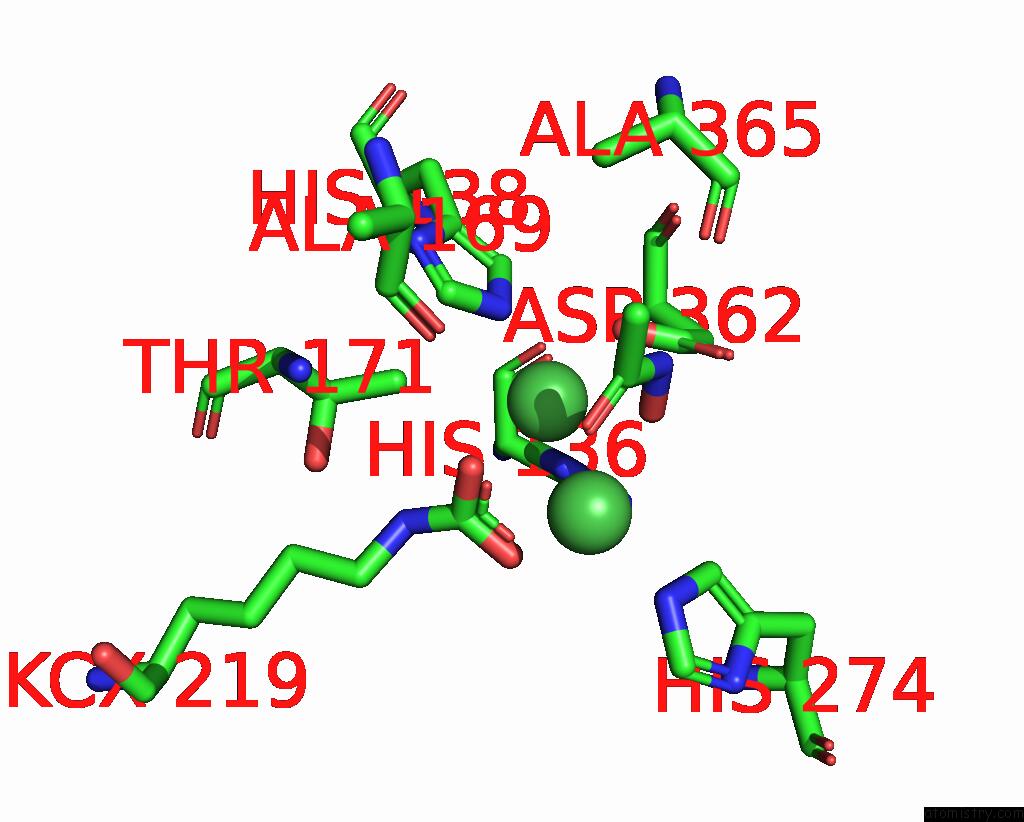
Mono view
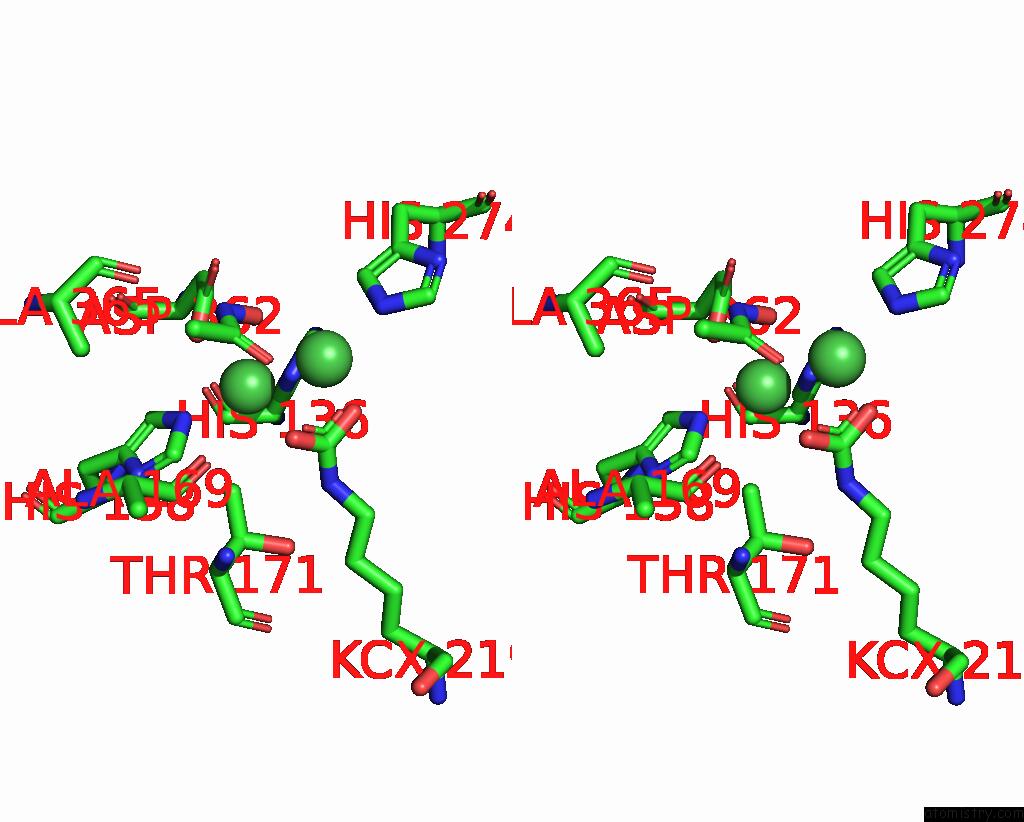
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Crystal Structure of Helicobacter Pylori Urease in Complex with Acetohydroxamic Acid within 5.0Å range:
|
Reference:
N.-C.Ha,
S.-T.Oh,
J.Y.Sung,
K.-A.Cha,
M.Hyung Lee,
B.-H.Oh.
Supramolecular Assembly and Acid Resistance of Helicobacter Pylori Urease Nat.Struct.Biol. V. 8 480 2001.
ISSN: ISSN 1072-8368
PubMed: 11373617
DOI: 10.1038/88563
Page generated: Mon Aug 18 17:38:00 2025
ISSN: ISSN 1072-8368
PubMed: 11373617
DOI: 10.1038/88563
Last articles
Ni in 5KE1Ni in 5K4L
Ni in 5K7H
Ni in 5JT1
Ni in 5JSK
Ni in 5JRD
Ni in 5JSU
Ni in 5JSH
Ni in 5JPT
Ni in 5JLX