Nickel »
PDB 4d00-4gu7 »
4dvo »
Nickel in PDB 4dvo: Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9
Enzymatic activity of Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9
All present enzymatic activity of Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9:
5.3.1.5;
5.3.1.5;
Protein crystallography data
The structure of Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9, PDB code: 4dvo
was solved by
A.Y.Kovalevsky,
L.Hanson,
P.Langan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.00 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.061, 99.428, 102.798, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19 / 21.4 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9
(pdb code 4dvo). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 3 binding sites of Nickel where determined in the Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9, PDB code: 4dvo:
Jump to Nickel binding site number: 1; 2; 3;
In total 3 binding sites of Nickel where determined in the Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9, PDB code: 4dvo:
Jump to Nickel binding site number: 1; 2; 3;
Nickel binding site 1 out of 3 in 4dvo
Go back to
Nickel binding site 1 out
of 3 in the Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9
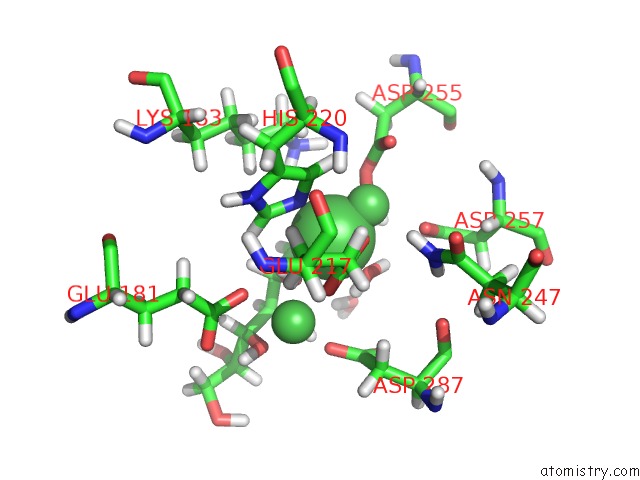
Mono view
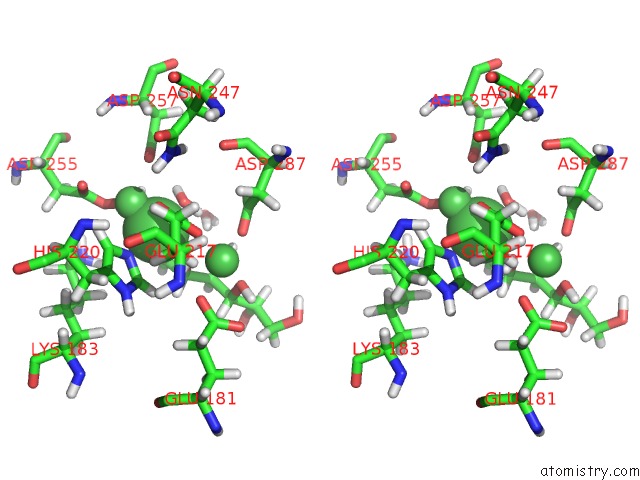
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9 within 5.0Å range:
|
Nickel binding site 2 out of 3 in 4dvo
Go back to
Nickel binding site 2 out
of 3 in the Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9
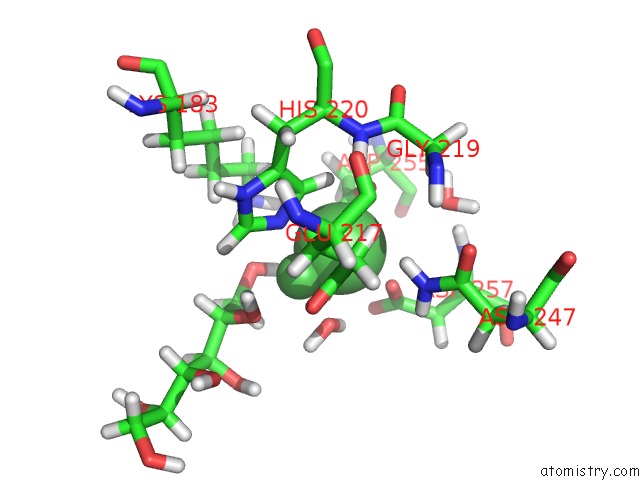
Mono view
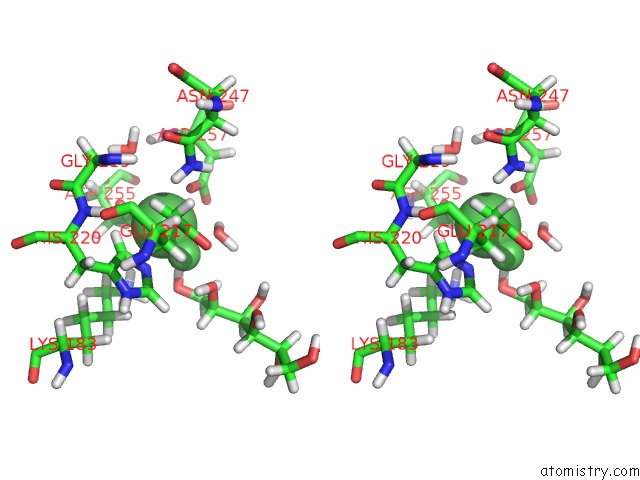
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9 within 5.0Å range:
|
Nickel binding site 3 out of 3 in 4dvo
Go back to
Nickel binding site 3 out
of 3 in the Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9
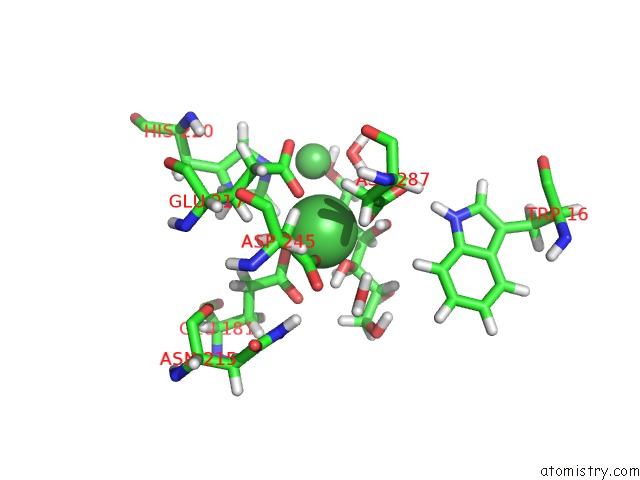
Mono view
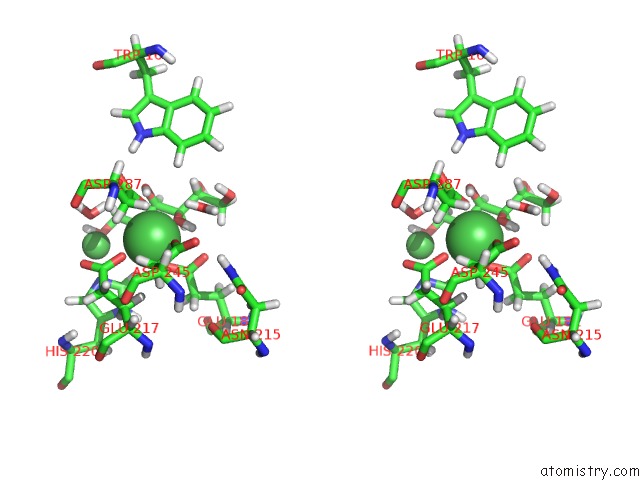
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 3 of Room-Temperature Joint X-Ray/Neutron Structure of D-Xylose Isomerase in Complex with 2NI2+ and Per-Deuterated D-Sorbitol at pH 5.9 within 5.0Å range:
|
Reference:
A.Kovalevsky,
B.L.Hanson,
S.A.Mason,
V.T.Forsyth,
Z.Fisher,
M.Mustyakimov,
M.P.Blakeley,
D.A.Keen,
P.Langan.
Inhibition of D-Xylose Isomerase By Polyols: Atomic Details By Joint X-Ray/Neutron Crystallography. Acta Crystallogr.,Sect.D V. 68 1201 2012.
ISSN: ISSN 0907-4449
PubMed: 22948921
DOI: 10.1107/S0907444912024808
Page generated: Mon Aug 18 19:11:18 2025
ISSN: ISSN 0907-4449
PubMed: 22948921
DOI: 10.1107/S0907444912024808
Last articles
Zn in 2K2DZn in 2K1Q
Zn in 2K1P
Zn in 2K1J
Zn in 2K17
Zn in 2K16
Zn in 2K0A
Zn in 2JZW
Zn in 2K0C
Zn in 2JYI