Nickel »
PDB 4d00-4gu7 »
4er8 »
Nickel in PDB 4er8: Structure of the Rep Associates Tyrosine Transposase Bound to A Rep Hairpin
Protein crystallography data
The structure of Structure of the Rep Associates Tyrosine Transposase Bound to A Rep Hairpin, PDB code: 4er8
was solved by
S.A.J.Messing,
B.Ton-Hoang,
A.B.Hickman,
R.Ghirlando,
M.Chandler,
F.Dyda,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 23.26 / 2.60 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 100.830, 60.300, 70.780, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.5 / 28.1 |
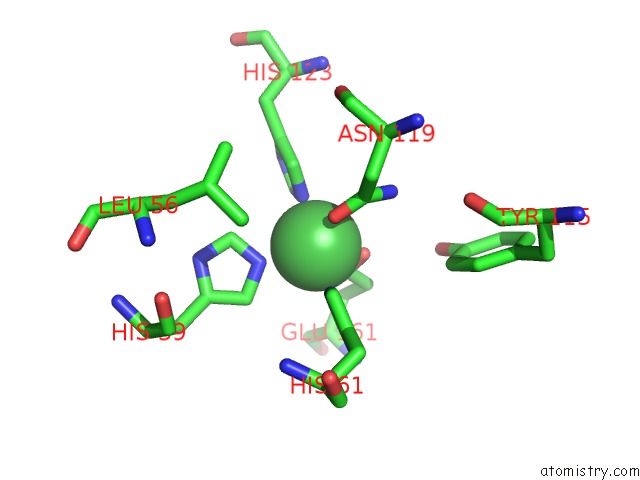
Nickel Binding Sites:
The binding sites of Nickel atom in the Structure of the Rep Associates Tyrosine Transposase Bound to A Rep Hairpin
(pdb code 4er8). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total only one binding site of Nickel was determined in the Structure of the Rep Associates Tyrosine Transposase Bound to A Rep Hairpin, PDB code: 4er8:
In total only one binding site of Nickel was determined in the Structure of the Rep Associates Tyrosine Transposase Bound to A Rep Hairpin, PDB code: 4er8:
Nickel binding site 1 out of 1 in 4er8
Go back to
Nickel binding site 1 out
of 1 in the Structure of the Rep Associates Tyrosine Transposase Bound to A Rep Hairpin
Mono view
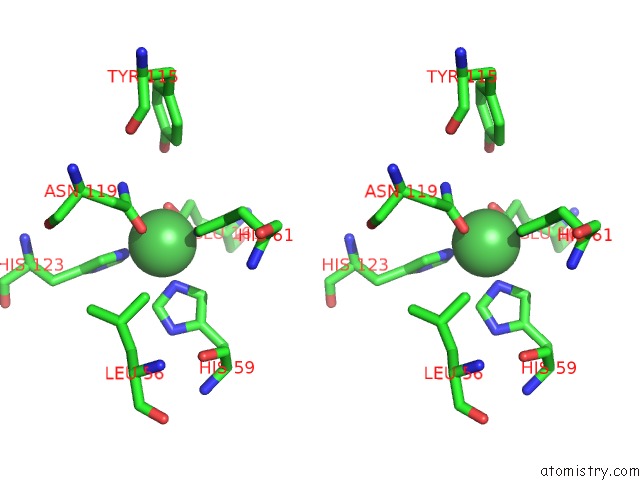
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Structure of the Rep Associates Tyrosine Transposase Bound to A Rep Hairpin within 5.0Å range:
|
Reference:
S.A.Messing,
B.Ton-Hoang,
A.B.Hickman,
A.J.Mccubbin,
G.F.Peaslee,
R.Ghirlando,
M.Chandler,
F.Dyda.
The Processing of Repetitive Extragenic Palindromes: the Structure of A Repetitive Extragenic Palindrome Bound to Its Associated Nuclease. Nucleic Acids Res. V. 40 9964 2012.
ISSN: ISSN 0305-1048
PubMed: 22885300
DOI: 10.1093/NAR/GKS741
Page generated: Mon Aug 18 19:12:12 2025
ISSN: ISSN 0305-1048
PubMed: 22885300
DOI: 10.1093/NAR/GKS741
Last articles
Fe in 9VR0Fe in 9UD8
Fe in 9QDT
Fe in 9S2T
Fe in 9JQA
Fe in 9IYV
Fe in 9J47
Fe in 9JQM
Fe in 9J2L
Fe in 9J1Q