Nickel »
PDB 4d00-4gu7 »
4eyu »
Nickel in PDB 4eyu: The Free Structure of the Mouse C-Terminal Domain of KDM6B
Protein crystallography data
The structure of The Free Structure of the Mouse C-Terminal Domain of KDM6B, PDB code: 4eyu
was solved by
Z.J.Cheng,
D.J.Patel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 38.45 / 2.30 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 82.977, 102.284, 145.302, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23 / 26.8 |
Other elements in 4eyu:
The structure of The Free Structure of the Mouse C-Terminal Domain of KDM6B also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Nickel Binding Sites:
The binding sites of Nickel atom in the The Free Structure of the Mouse C-Terminal Domain of KDM6B
(pdb code 4eyu). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the The Free Structure of the Mouse C-Terminal Domain of KDM6B, PDB code: 4eyu:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the The Free Structure of the Mouse C-Terminal Domain of KDM6B, PDB code: 4eyu:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 4eyu
Go back to
Nickel binding site 1 out
of 2 in the The Free Structure of the Mouse C-Terminal Domain of KDM6B
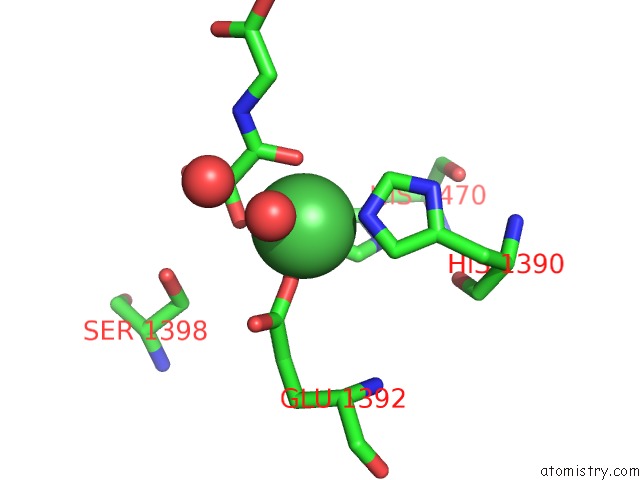
Mono view
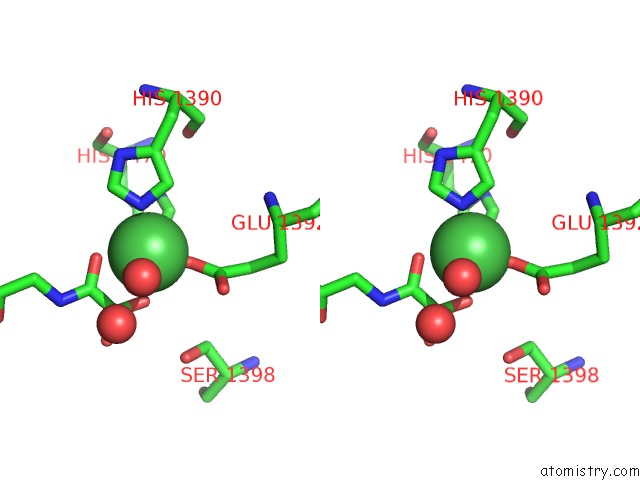
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of The Free Structure of the Mouse C-Terminal Domain of KDM6B within 5.0Å range:
|
Nickel binding site 2 out of 2 in 4eyu
Go back to
Nickel binding site 2 out
of 2 in the The Free Structure of the Mouse C-Terminal Domain of KDM6B
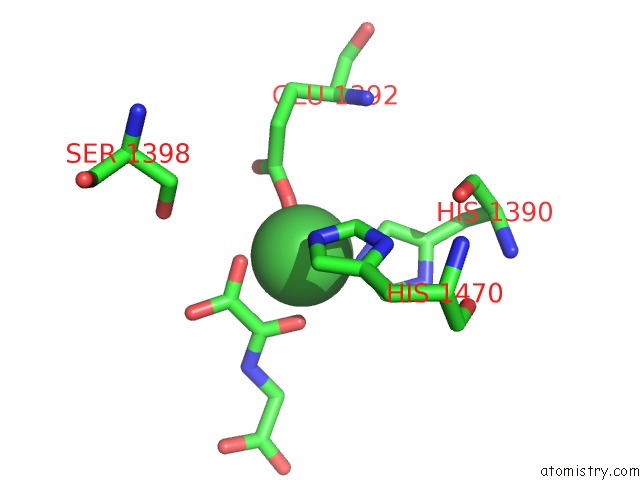
Mono view
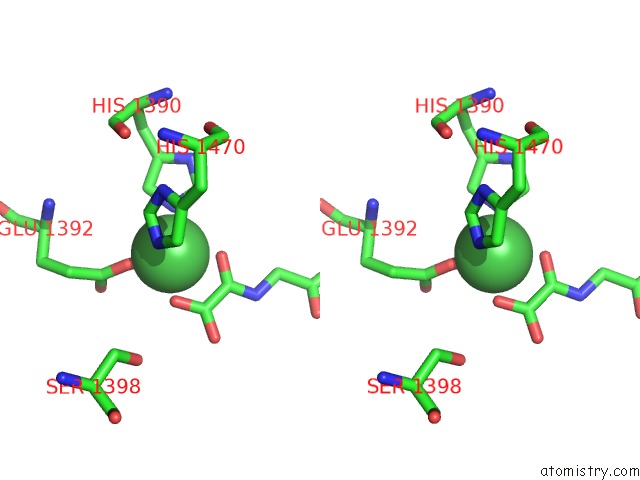
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of The Free Structure of the Mouse C-Terminal Domain of KDM6B within 5.0Å range:
|
Reference:
L.Kruidenier,
C.W.Chung,
Z.Cheng,
J.Liddle,
K.Che,
G.Joberty,
M.Bantscheff,
C.Bountra,
A.Bridges,
H.Diallo,
D.Eberhard,
S.Hutchinson,
E.Jones,
R.Katso,
M.Leveridge,
P.K.Mander,
J.Mosley,
C.Ramirez-Molina,
P.Rowland,
C.J.Schofield,
R.J.Sheppard,
J.E.Smith,
C.Swales,
R.Tanner,
P.Thomas,
A.Tumber,
G.Drewes,
U.Oppermann,
D.J.Patel,
K.Lee,
D.M.Wilson.
A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response. Nature V. 488 404 2012.
ISSN: ISSN 0028-0836
PubMed: 22842901
DOI: 10.1038/NATURE11262
Page generated: Mon Aug 18 19:12:39 2025
ISSN: ISSN 0028-0836
PubMed: 22842901
DOI: 10.1038/NATURE11262
Last articles
Ni in 5U7HNi in 5V2P
Ni in 5U9E
Ni in 5TVS
Ni in 5U93
Ni in 5TVR
Ni in 5TRQ
Ni in 5TFZ
Ni in 5TR8
Ni in 5T22