Nickel »
PDB 4ofo-4rro »
4omo »
Nickel in PDB 4omo: Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E
Enzymatic activity of Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E
All present enzymatic activity of Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E:
2.7.10.2;
2.7.10.2;
Protein crystallography data
The structure of Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E, PDB code: 4omo
was solved by
A.Camara-Artigas,
J.Bacarizo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.11 / 1.04 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.711, 45.464, 57.807, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.8 / 16.8 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E
(pdb code 4omo). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E, PDB code: 4omo:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E, PDB code: 4omo:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 4omo
Go back to
Nickel binding site 1 out
of 2 in the Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E
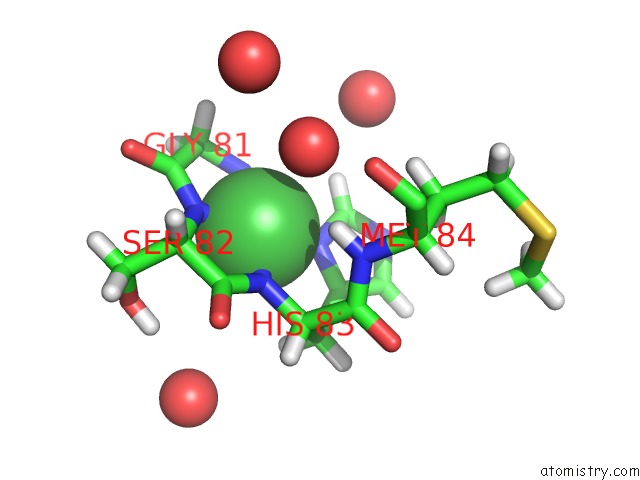
Mono view
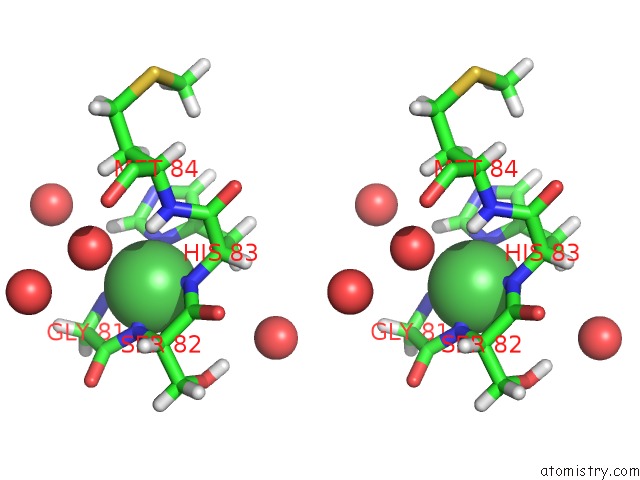
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E within 5.0Å range:
|
Nickel binding site 2 out of 2 in 4omo
Go back to
Nickel binding site 2 out
of 2 in the Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E
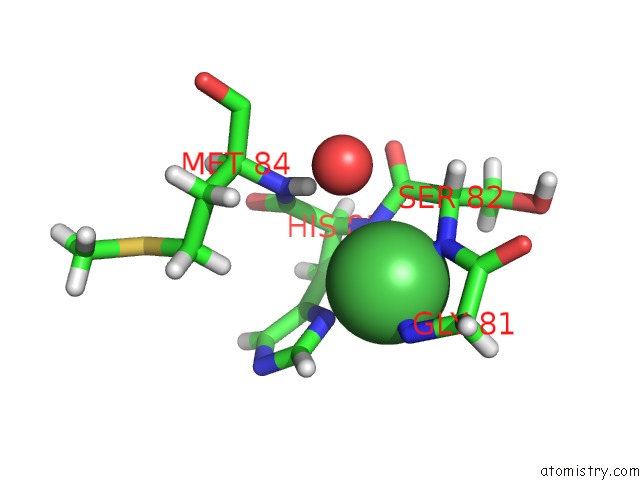
Mono view
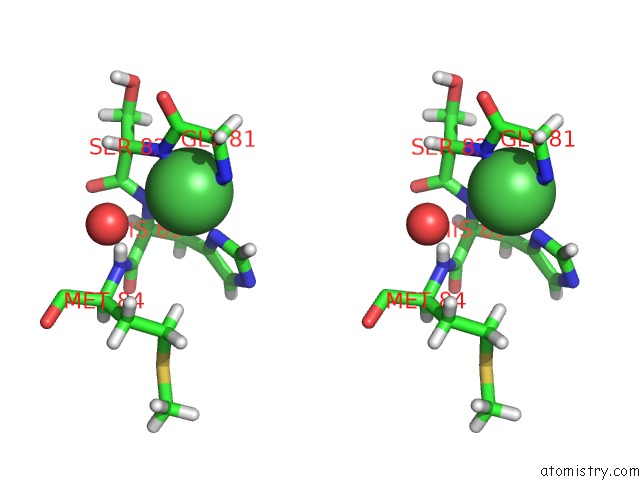
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Crystal Structure of the C-Src Tyrosine Kinase SH3 Domain Mutant Q128E within 5.0Å range:
|
Reference:
J.Bacarizo,
S.Martinez-Rodriguez,
J.M.Martin-Garcia,
M.Andujar-Sanchez,
E.Ortiz-Salmeron,
J.L.Neira,
A.Camara-Artigas.
Electrostatic Effects in the Folding of the SH3 Domain of the C-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation. Plos One V. 9 13224 2014.
ISSN: ESSN 1932-6203
PubMed: 25490095
DOI: 10.1371/JOURNAL.PONE.0113224
Page generated: Mon Aug 18 19:35:25 2025
ISSN: ESSN 1932-6203
PubMed: 25490095
DOI: 10.1371/JOURNAL.PONE.0113224
Last articles
Pt in 6AQKPt in 6BS1
Pt in 6BRX
Pt in 5Z80
Pt in 5YKY
Pt in 5V6I
Pt in 5T7L
Pt in 5NJ7
Pt in 5OLD
Pt in 5OLE