Nickel »
PDB 4ofo-4rro »
4q2d »
Nickel in PDB 4q2d: Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate
Protein crystallography data
The structure of Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate, PDB code: 4q2d
was solved by
B.Gong,
M.Shin,
J.Sun,
J.Van Der Oost,
J.-S.Kim,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.66 / 2.77 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 105.684, 211.630, 104.272, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.7 / 24.7 |
Other elements in 4q2d:
The structure of Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Nickel Binding Sites:
The binding sites of Nickel atom in the Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate
(pdb code 4q2d). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 3 binding sites of Nickel where determined in the Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate, PDB code: 4q2d:
Jump to Nickel binding site number: 1; 2; 3;
In total 3 binding sites of Nickel where determined in the Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate, PDB code: 4q2d:
Jump to Nickel binding site number: 1; 2; 3;
Nickel binding site 1 out of 3 in 4q2d
Go back to
Nickel binding site 1 out
of 3 in the Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate
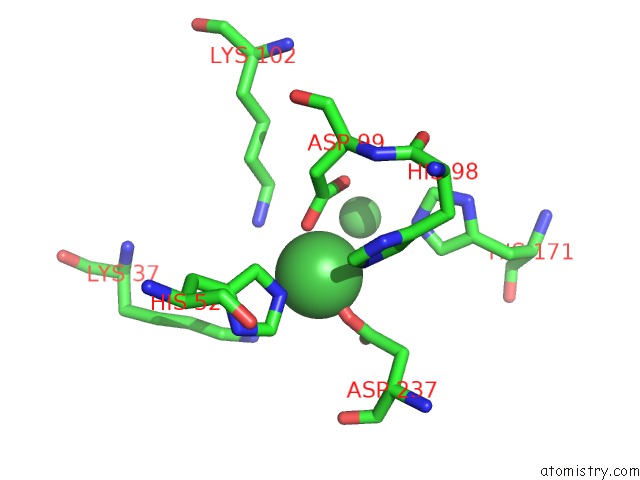
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate within 5.0Å range:
|
Nickel binding site 2 out of 3 in 4q2d
Go back to
Nickel binding site 2 out
of 3 in the Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate within 5.0Å range:
|
Nickel binding site 3 out of 3 in 4q2d
Go back to
Nickel binding site 3 out
of 3 in the Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 3 of Crystal Structure of Crispr-Associated Protein in Complex with 2'- Deoxyadenosine 5'-Triphosphate within 5.0Å range:
|
Reference:
B.Gong,
M.Shin,
J.Sun,
C.H.Jung,
E.L.Bolt,
J.Van Der Oost,
J.-S.Kim.
Molecular Insights Into Dna Interference By Crispr-Associated Nuclease-Helicase CAS3 Proc.Natl.Acad.Sci.Usa 2014.
ISSN: ESSN 1091-6490
PubMed: 25368186
DOI: 10.1073/PNAS.1410806111
Page generated: Mon Aug 18 19:36:32 2025
ISSN: ESSN 1091-6490
PubMed: 25368186
DOI: 10.1073/PNAS.1410806111
Last articles
Rb in 6DOTRb in 5YLV
Rb in 5AW8
Rb in 5YCK
Rb in 5AW6
Rb in 5AW4
Rb in 5AW7
Rb in 5AW5
Rb in 4E8P
Rb in 4R8C