Nickel »
PDB 4ofo-4rro »
4qdw »
Nickel in PDB 4qdw: Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose
Enzymatic activity of Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose
All present enzymatic activity of Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose:
5.3.1.5;
5.3.1.5;
Protein crystallography data
The structure of Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose, PDB code: 4qdw
was solved by
A.Y.Kovalevsky,
P.Langan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 1.80 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.190, 99.670, 102.940, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.6 / 17.9 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose
(pdb code 4qdw). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 3 binding sites of Nickel where determined in the Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose, PDB code: 4qdw:
Jump to Nickel binding site number: 1; 2; 3;
In total 3 binding sites of Nickel where determined in the Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose, PDB code: 4qdw:
Jump to Nickel binding site number: 1; 2; 3;
Nickel binding site 1 out of 3 in 4qdw
Go back to
Nickel binding site 1 out
of 3 in the Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose
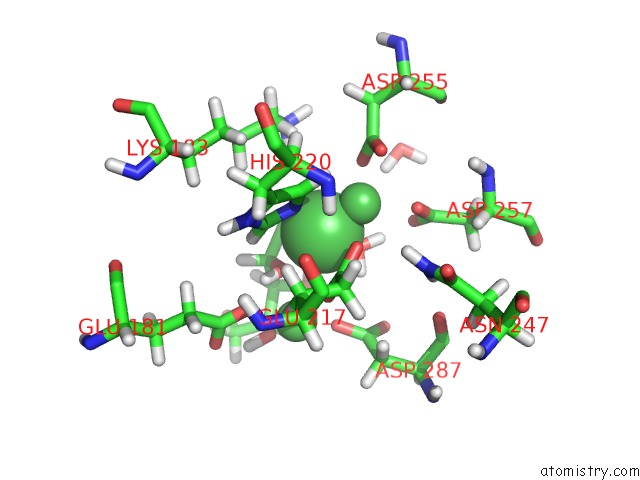
Mono view
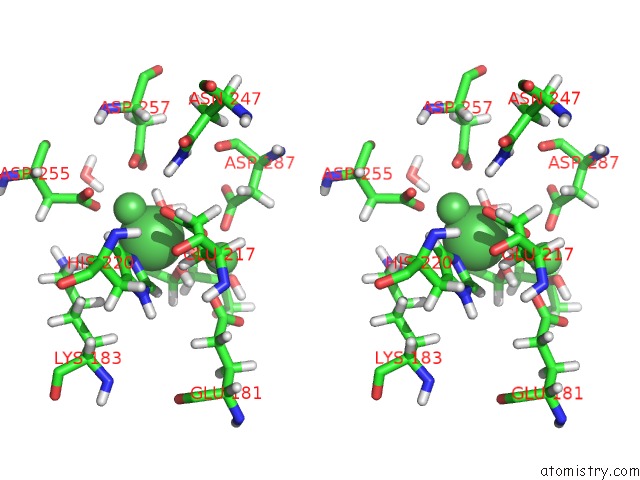
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose within 5.0Å range:
|
Nickel binding site 2 out of 3 in 4qdw
Go back to
Nickel binding site 2 out
of 3 in the Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose
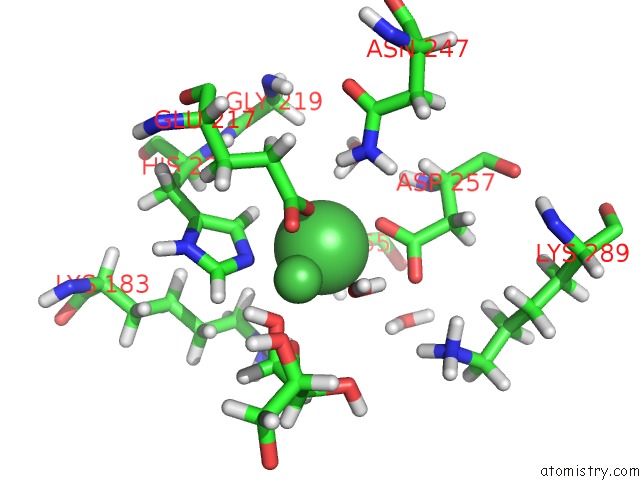
Mono view
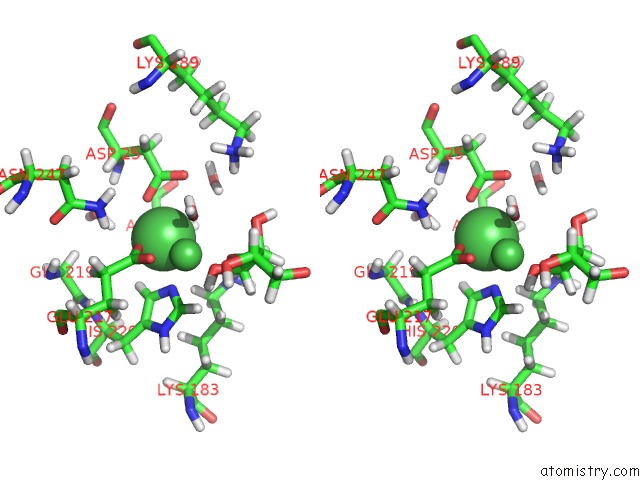
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose within 5.0Å range:
|
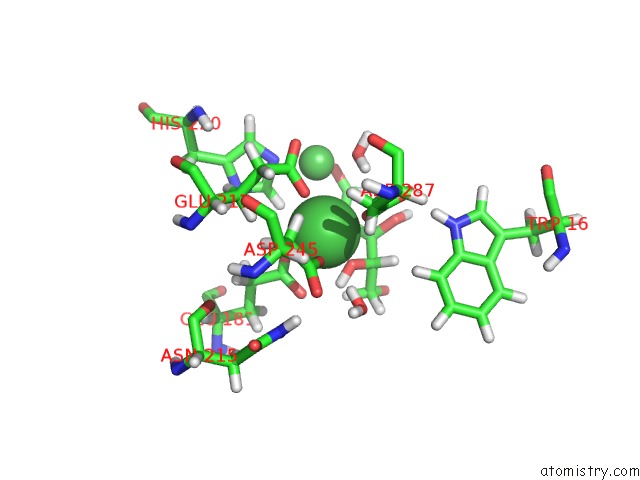
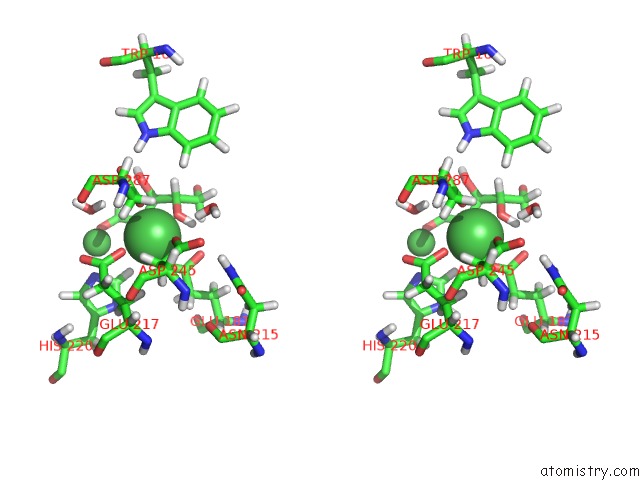
Nickel binding site 3 out of 3 in 4qdw
Go back to
Nickel binding site 3 out
of 3 in the Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 3 of Joint X-Ray and Neutron Structure of Streptomyces Rubiginosus D-Xylose Isomerase in Complex with Two NI2+ Ions and Linear L-Arabinose within 5.0Å range:
|
Reference:
P.Langan,
A.K.Sangha,
T.Wymore,
J.M.Parks,
Z.K.Yang,
B.L.Hanson,
Z.Fisher,
S.A.Mason,
M.P.Blakeley,
V.T.Forsyth,
J.P.Glusker,
H.L.Carrell,
J.C.Smith,
D.A.Keen,
D.E.Graham,
A.Kovalevsky.
L-Arabinose Binding, Isomerization, and Epimerization By D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study. Structure V. 22 1287 2014.
ISSN: ISSN 0969-2126
PubMed: 25132082
DOI: 10.1016/J.STR.2014.07.002
Page generated: Mon Aug 18 19:37:19 2025
ISSN: ISSN 0969-2126
PubMed: 25132082
DOI: 10.1016/J.STR.2014.07.002
Last articles
K in 9NESK in 9PHG
K in 9NEI
K in 9NED
K in 9NEC
K in 9NEG
K in 9CWU
K in 9CVB
K in 9CVA
K in 9COM