Nickel »
PDB 4ofo-4rro »
4qx8 »
Nickel in PDB 4qx8: Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg
Enzymatic activity of Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg
All present enzymatic activity of Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg:
1.14.11.27;
1.14.11.27;
Protein crystallography data
The structure of Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg, PDB code: 4qx8
was solved by
Z.J.Cheng,
D.J Patel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 85.04 / 1.65 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 53.797, 84.615, 170.077, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.8 / 22.7 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg
(pdb code 4qx8). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg, PDB code: 4qx8:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg, PDB code: 4qx8:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 4qx8
Go back to
Nickel binding site 1 out
of 2 in the Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg
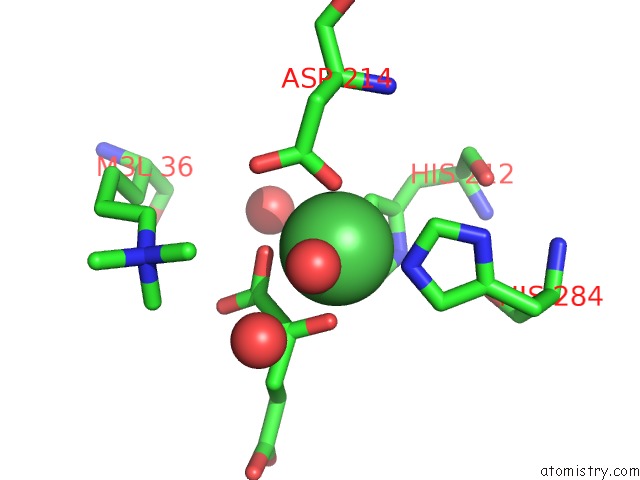
Mono view
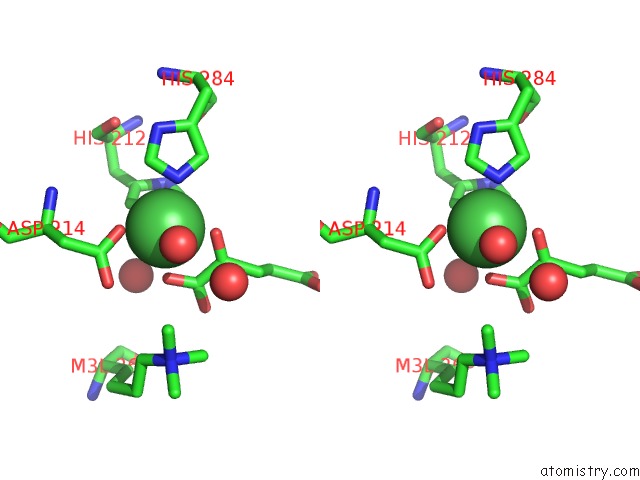
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg within 5.0Å range:
|
Nickel binding site 2 out of 2 in 4qx8
Go back to
Nickel binding site 2 out
of 2 in the Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg
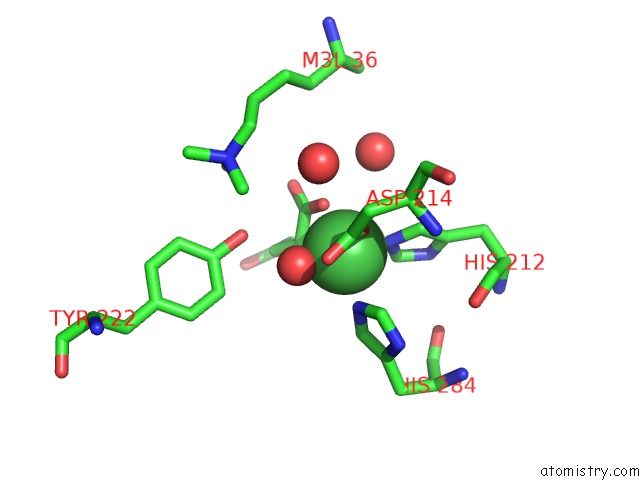
Mono view
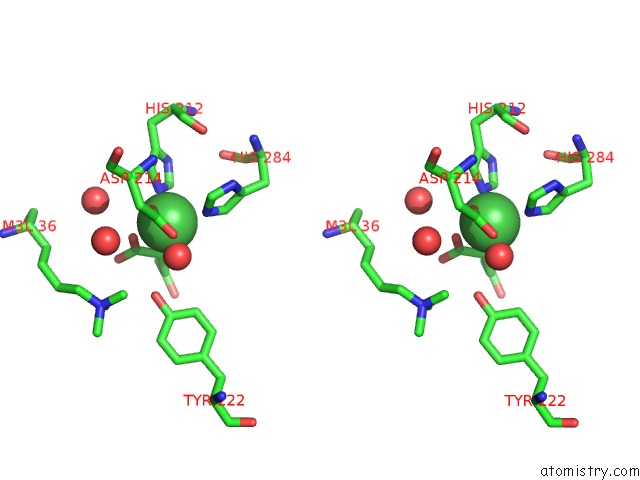
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Crystal Structure of Histone Demethylase KDM2A-H3K36ME3 Complex with Alpha-Kg within 5.0Å range:
|
Reference:
Z.Cheng,
P.Cheung,
A.J.Kuo,
E.T.Yukl,
C.M.Wilmot,
O.Gozani,
D.J.Patel.
A Molecular Threading Mechanism Underlies Jumonji Lysine Demethylase KDM2A Regulation of Methylated H3K36. Genes Dev. V. 28 1758 2014.
ISSN: ISSN 0890-9369
PubMed: 25128496
DOI: 10.1101/GAD.246561.114
Page generated: Mon Aug 18 19:38:28 2025
ISSN: ISSN 0890-9369
PubMed: 25128496
DOI: 10.1101/GAD.246561.114
Last articles
Ni in 7W1FNi in 7VXQ
Ni in 7W58
Ni in 7VUD
Ni in 7VQ6
Ni in 7VTB
Ni in 7VGB
Ni in 7UUR
Ni in 7VJS
Ni in 7VEI