Nickel »
PDB 5e6m-5i14 »
5fsd »
Nickel in PDB 5fsd: 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease
Enzymatic activity of 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease
All present enzymatic activity of 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease:
3.5.1.5;
3.5.1.5;
Protein crystallography data
The structure of 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease, PDB code: 5fsd
was solved by
L.Mazzei,
M.Cianci,
F.Musiani,
S.Ciurli,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 113.72 / 1.75 |
| Space group | P 63 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 131.312, 131.312, 188.920, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 13.4 / 17 |
Nickel Binding Sites:
The binding sites of Nickel atom in the 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease
(pdb code 5fsd). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease, PDB code: 5fsd:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease, PDB code: 5fsd:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 5fsd
Go back to
Nickel binding site 1 out
of 2 in the 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease
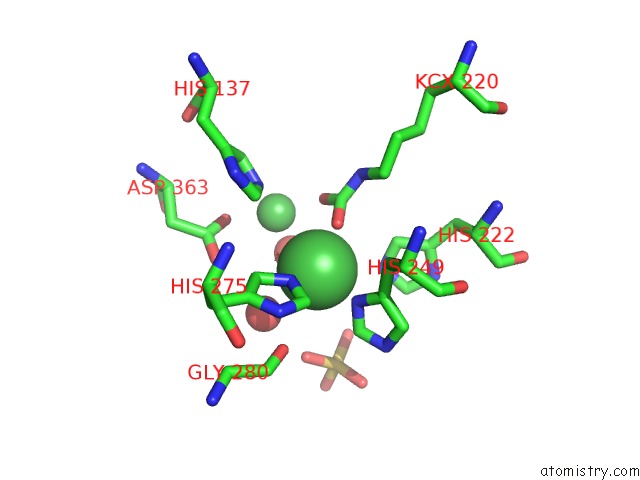
Mono view
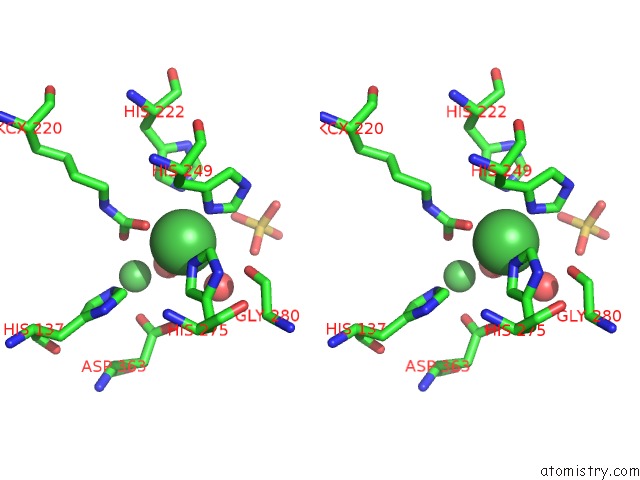
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease within 5.0Å range:
|
Nickel binding site 2 out of 2 in 5fsd
Go back to
Nickel binding site 2 out
of 2 in the 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease
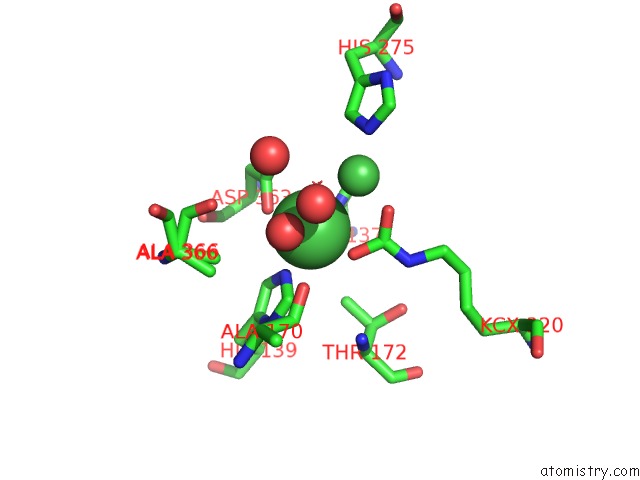
Mono view
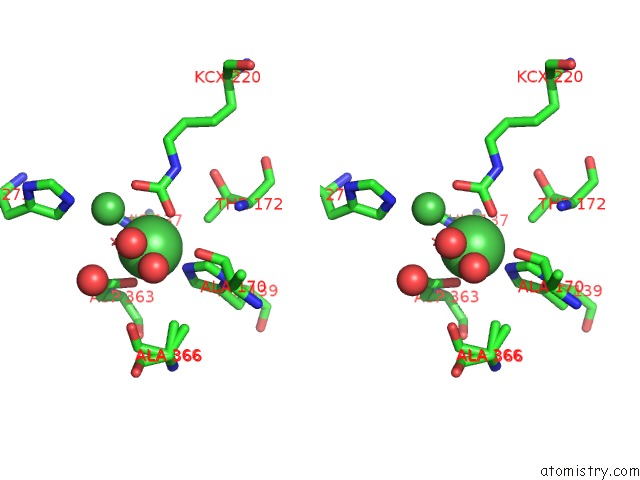
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of 1.75 A Resolution 2,5-Dihydroxybenzensulfonate Inhibited Sporosarcina Pasteurii Urease within 5.0Å range:
|
Reference:
L.Mazzei,
M.Cianci,
F.Musiani,
S.Ciurli.
Inactivation of Urease By 1,4-Benzoquinone: Chemistry at the Protein Surface. Dalton Trans V. 45 5455 2016.
ISSN: ISSN 1477-9226
PubMed: 26961812
DOI: 10.1039/C6DT00652C
Page generated: Mon Aug 18 20:04:35 2025
ISSN: ISSN 1477-9226
PubMed: 26961812
DOI: 10.1039/C6DT00652C
Last articles
Pb in 6A85Pb in 6EGP
Pb in 5XM8
Pb in 5VFF
Pb in 5VFG
Pb in 5VFE
Pb in 4XJN
Pb in 5U88
Pb in 5U7C
Pb in 5GPE