Nickel »
PDB 5q5u-5q6x »
5q60 »
Nickel in PDB 5q60: Pandda Analysis Group Deposition -- Crystal Structure of DCLRE1A After Initial Refinement with No Ligand Modelled (Structure 137)
Protein crystallography data
The structure of Pandda Analysis Group Deposition -- Crystal Structure of DCLRE1A After Initial Refinement with No Ligand Modelled (Structure 137), PDB code: 5q60
was solved by
J.A.Newman,
H.Aitkenhead,
S.Y.Lee,
K.Kupinska,
N.Burgess-Brown,
R.Tallon,
T.Krojer,
F.Von Delft,
C.H.Arrowsmith,
A.Edwards,
C.Bountra,
O.Gileadi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 51.97 / 1.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.972, 57.220, 115.309, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.6 / 23.9 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Pandda Analysis Group Deposition -- Crystal Structure of DCLRE1A After Initial Refinement with No Ligand Modelled (Structure 137)
(pdb code 5q60). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total only one binding site of Nickel was determined in the Pandda Analysis Group Deposition -- Crystal Structure of DCLRE1A After Initial Refinement with No Ligand Modelled (Structure 137), PDB code: 5q60:
In total only one binding site of Nickel was determined in the Pandda Analysis Group Deposition -- Crystal Structure of DCLRE1A After Initial Refinement with No Ligand Modelled (Structure 137), PDB code: 5q60:
Nickel binding site 1 out of 1 in 5q60
Go back to
Nickel binding site 1 out
of 1 in the Pandda Analysis Group Deposition -- Crystal Structure of DCLRE1A After Initial Refinement with No Ligand Modelled (Structure 137)
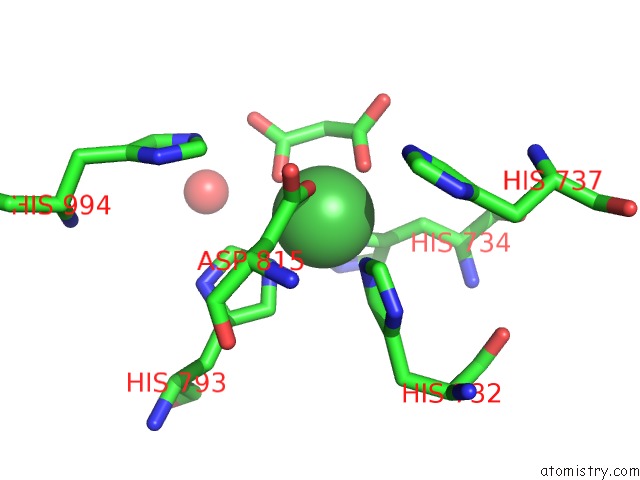
Mono view
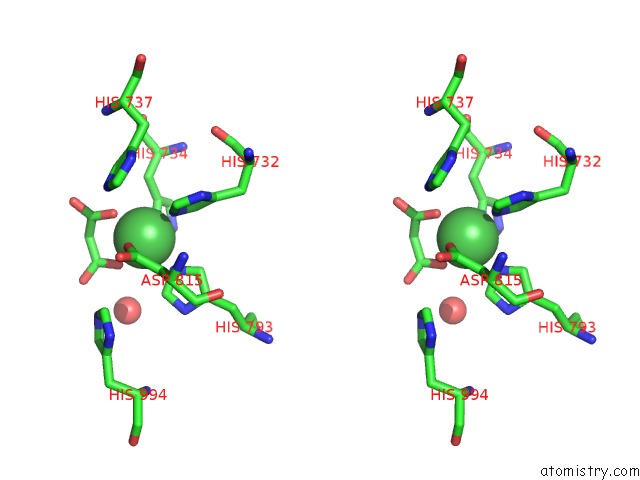
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Pandda Analysis Group Deposition -- Crystal Structure of DCLRE1A After Initial Refinement with No Ligand Modelled (Structure 137) within 5.0Å range:
|
Reference:
J.A.Newman,
H.Aitkenhead,
S.Y.Lee,
K.Kupinska,
N.Burgess-Brown,
R.Tallon,
T.Krojer,
F.Von Delft,
C.H.Arrowsmith,
A.Edwards,
C.Bountra,
O.Gileadi.
Pandda Analysis Group Deposition To Be Published.
Page generated: Mon Aug 18 20:44:38 2025
Last articles
Pt in 4QOTPt in 4R6C
Pt in 4Q8F
Pt in 4QGZ
Pt in 4Q8E
Pt in 4PKE
Pt in 4Q37
Pt in 4PM3
Pt in 4PGW
Pt in 4PPO