Nickel »
PDB 3dse-3hy3 »
3g50 »
Nickel in PDB 3g50: Crystal Structure of Nisod D3A Mutant at 1.9 A
Enzymatic activity of Crystal Structure of Nisod D3A Mutant at 1.9 A
All present enzymatic activity of Crystal Structure of Nisod D3A Mutant at 1.9 A:
1.15.1.1;
1.15.1.1;
Protein crystallography data
The structure of Crystal Structure of Nisod D3A Mutant at 1.9 A, PDB code: 3g50
was solved by
S.C.Garman,
A.I.Guce,
R.W.Herbst,
P.A.Bryngelson,
D.E.Cabelli,
K.A.Higgins,
K.C.Ryan,
M.J.Maroney,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 21.57 / 1.90 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.988, 112.274, 111.695, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.6 / 24 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Crystal Structure of Nisod D3A Mutant at 1.9 A
(pdb code 3g50). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the Crystal Structure of Nisod D3A Mutant at 1.9 A, PDB code: 3g50:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the Crystal Structure of Nisod D3A Mutant at 1.9 A, PDB code: 3g50:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 3g50
Go back to
Nickel binding site 1 out
of 2 in the Crystal Structure of Nisod D3A Mutant at 1.9 A
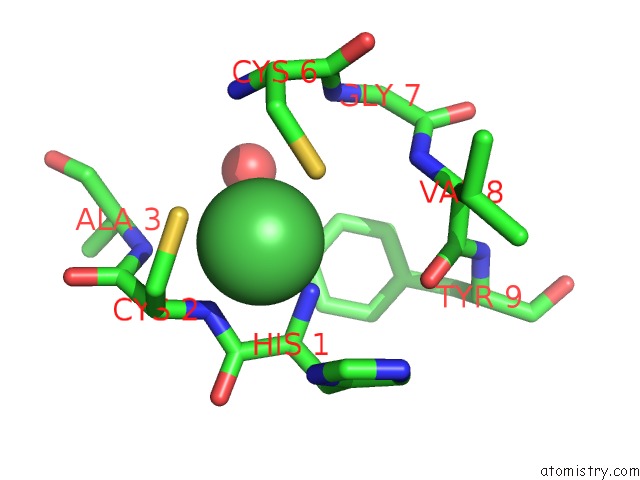
Mono view
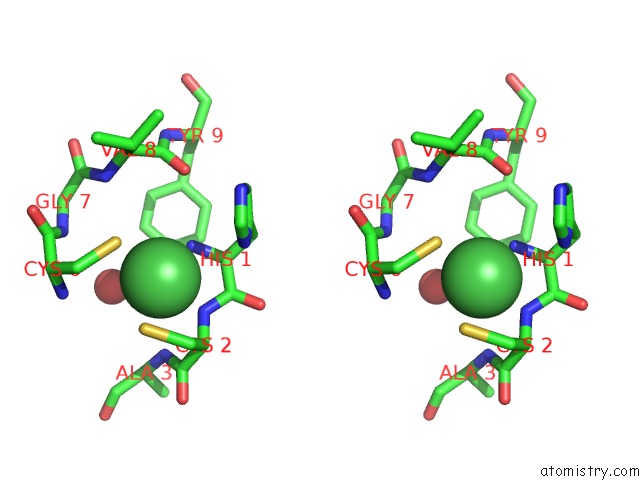
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Crystal Structure of Nisod D3A Mutant at 1.9 A within 5.0Å range:
|
Nickel binding site 2 out of 2 in 3g50
Go back to
Nickel binding site 2 out
of 2 in the Crystal Structure of Nisod D3A Mutant at 1.9 A
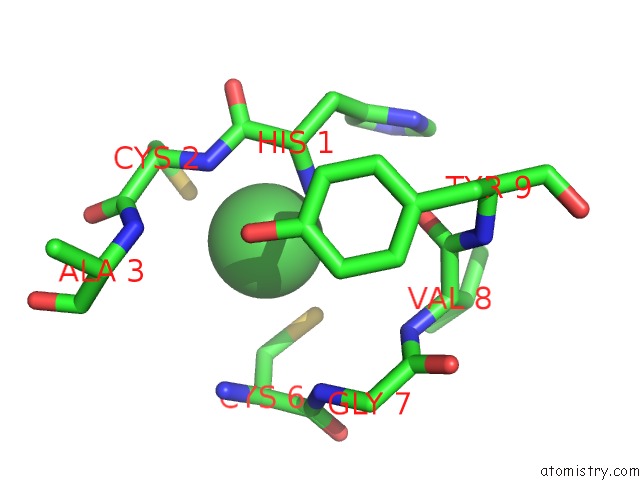
Mono view
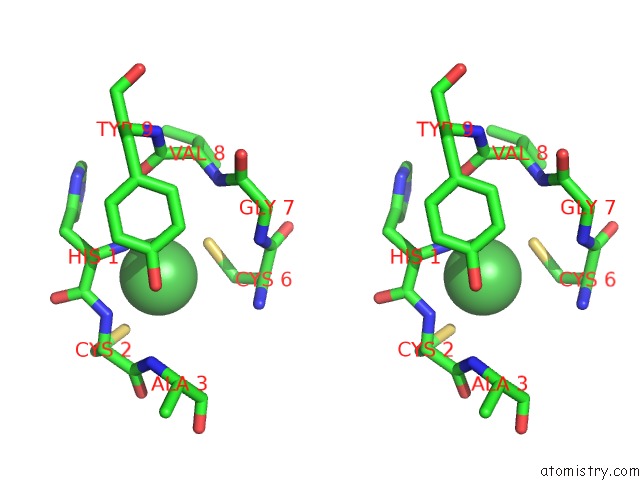
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Crystal Structure of Nisod D3A Mutant at 1.9 A within 5.0Å range:
|
Reference:
R.W.Herbst,
A.Guce,
P.A.Bryngelson,
K.A.Higgins,
K.C.Ryan,
D.E.Cabelli,
S.C.Garman,
M.J.Maroney.
Role of Conserved Tyrosine Residues in Nisod Catalysis: A Case of Convergent Evolution Biochemistry V. 48 3354 2009.
ISSN: ISSN 0006-2960
PubMed: 19183068
DOI: 10.1021/BI802029T
Page generated: Wed Oct 9 17:17:57 2024
ISSN: ISSN 0006-2960
PubMed: 19183068
DOI: 10.1021/BI802029T
Last articles
Mg in 5SDYMg in 5SDW
Mg in 5SDV
Mg in 5SDU
Mg in 5SCL
Mg in 5SCI
Mg in 5SCJ
Mg in 5SCK
Mg in 5SCG
Mg in 5SCH