Nickel »
PDB 3hy4-3kvb »
3idq »
Nickel in PDB 3idq: Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution
Enzymatic activity of Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution
All present enzymatic activity of Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution:
3.6.3.16;
3.6.3.16;
Protein crystallography data
The structure of Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution, PDB code: 3idq
was solved by
C.J.M.Suloway,
J.W.Chartron,
M.Zaslaver,
W.M.Clemons Jr.,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.05 / 3.70 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 115.320, 115.320, 281.111, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 28.3 / 33.5 |
Other elements in 3idq:
The structure of Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Nickel Binding Sites:
The binding sites of Nickel atom in the Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution
(pdb code 3idq). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total only one binding site of Nickel was determined in the Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution, PDB code: 3idq:
In total only one binding site of Nickel was determined in the Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution, PDB code: 3idq:
Nickel binding site 1 out of 1 in 3idq
Go back to
Nickel binding site 1 out
of 1 in the Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution
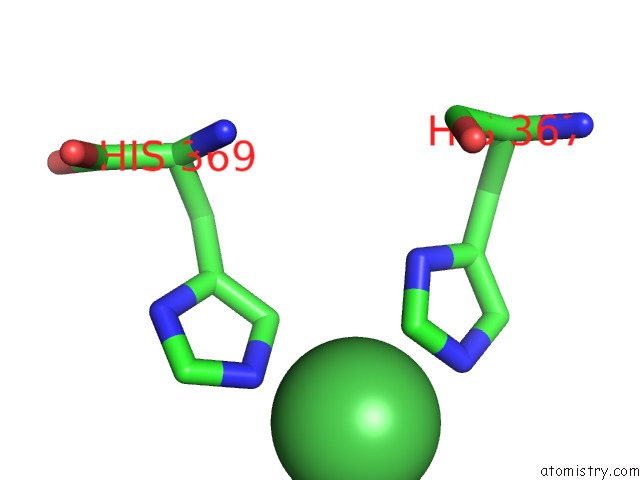
Mono view
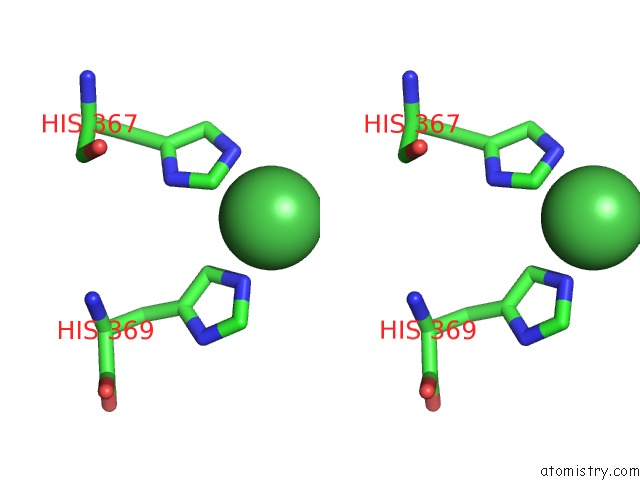
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Crystal Structure of S. Cerevisiae GET3 at 3.7 Angstrom Resolution within 5.0Å range:
|
Reference:
C.J.Suloway,
J.W.Chartron,
M.Zaslaver,
W.M.Clemons.
Model For Eukaryotic Tail-Anchored Protein Binding Based on the Structure of GET3 Proc.Natl.Acad.Sci.Usa V. 106 14849 2009.
ISSN: ISSN 0027-8424
PubMed: 19706470
DOI: 10.1073/PNAS.0907522106
Page generated: Wed Oct 9 17:23:27 2024
ISSN: ISSN 0027-8424
PubMed: 19706470
DOI: 10.1073/PNAS.0907522106
Last articles
Mg in 5CKIMg in 5CK3
Mg in 5CJV
Mg in 5CJY
Mg in 5CJN
Mg in 5CJU
Mg in 5CJ2
Mg in 5CJT
Mg in 5CJP
Mg in 5CJM