Nickel »
PDB 9gnr-9qm5 »
9gnr »
Nickel in PDB 9gnr: Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto
Enzymatic activity of Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto
All present enzymatic activity of Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto:
3.5.1.5;
3.5.1.5;
Nickel Binding Sites:
The binding sites of Nickel atom in the Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto
(pdb code 9gnr). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto, PDB code: 9gnr:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto, PDB code: 9gnr:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 9gnr
Go back to
Nickel binding site 1 out
of 2 in the Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto
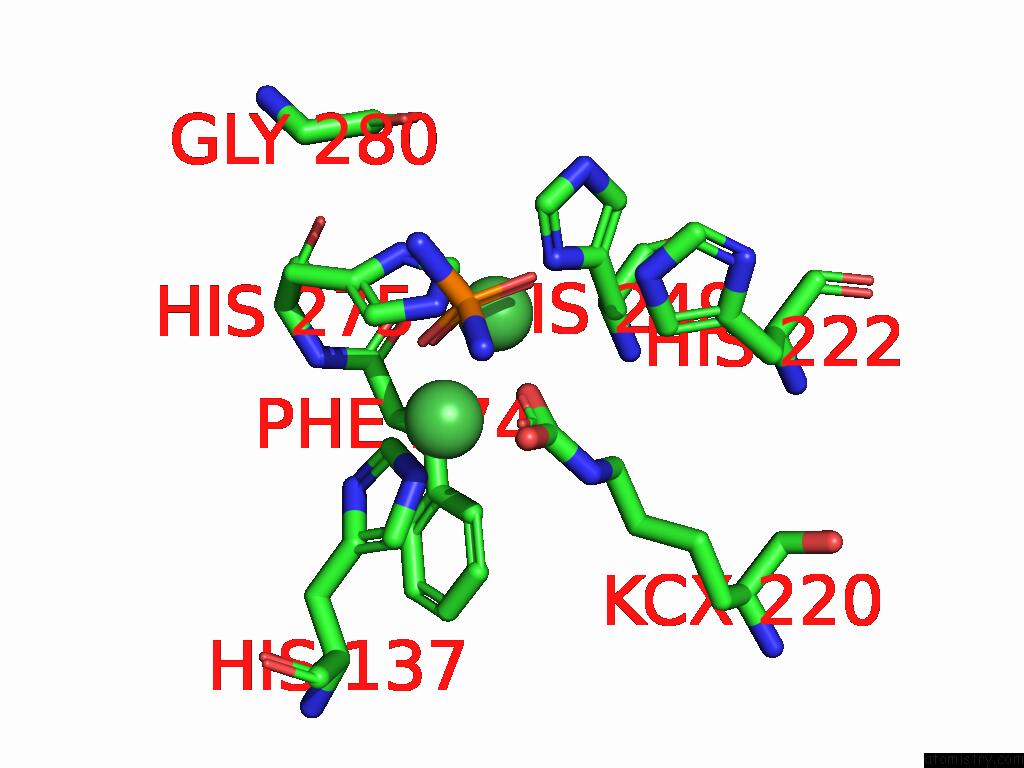
Mono view
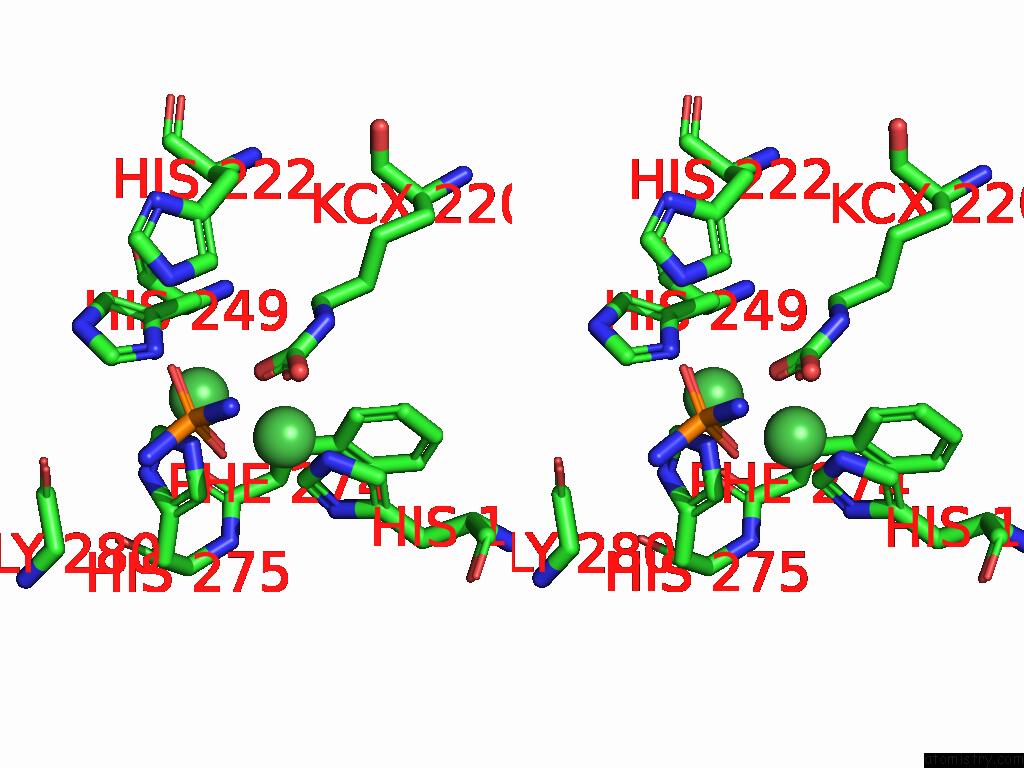
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto within 5.0Å range:
|
Nickel binding site 2 out of 2 in 9gnr
Go back to
Nickel binding site 2 out
of 2 in the Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto
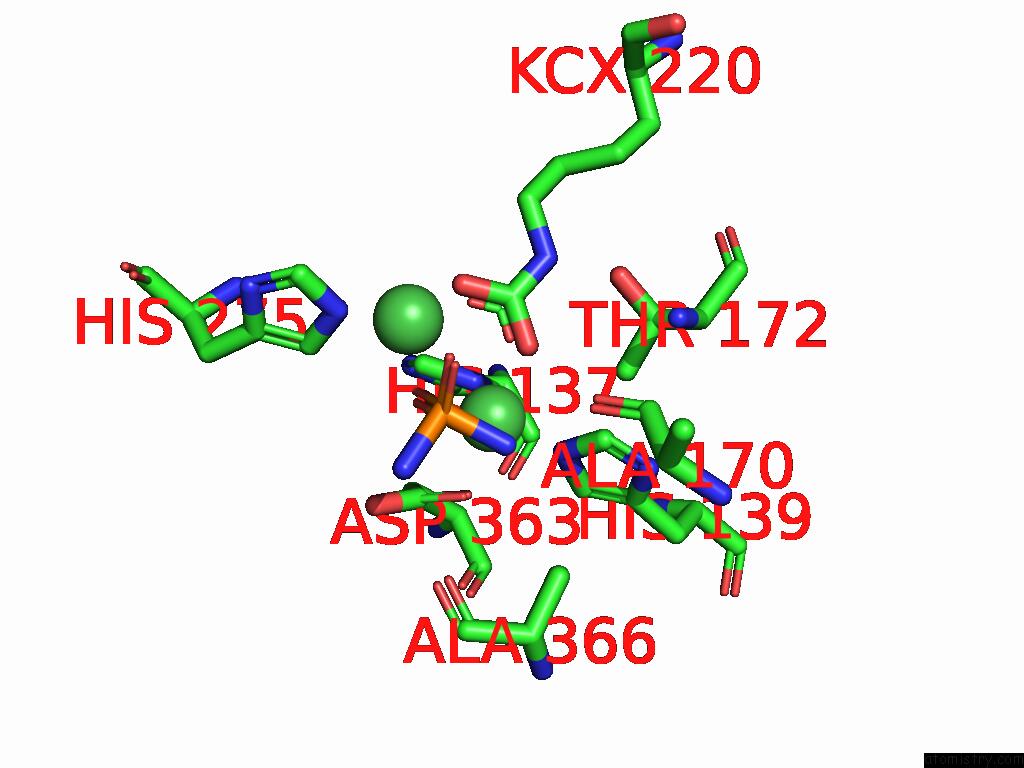
Mono view
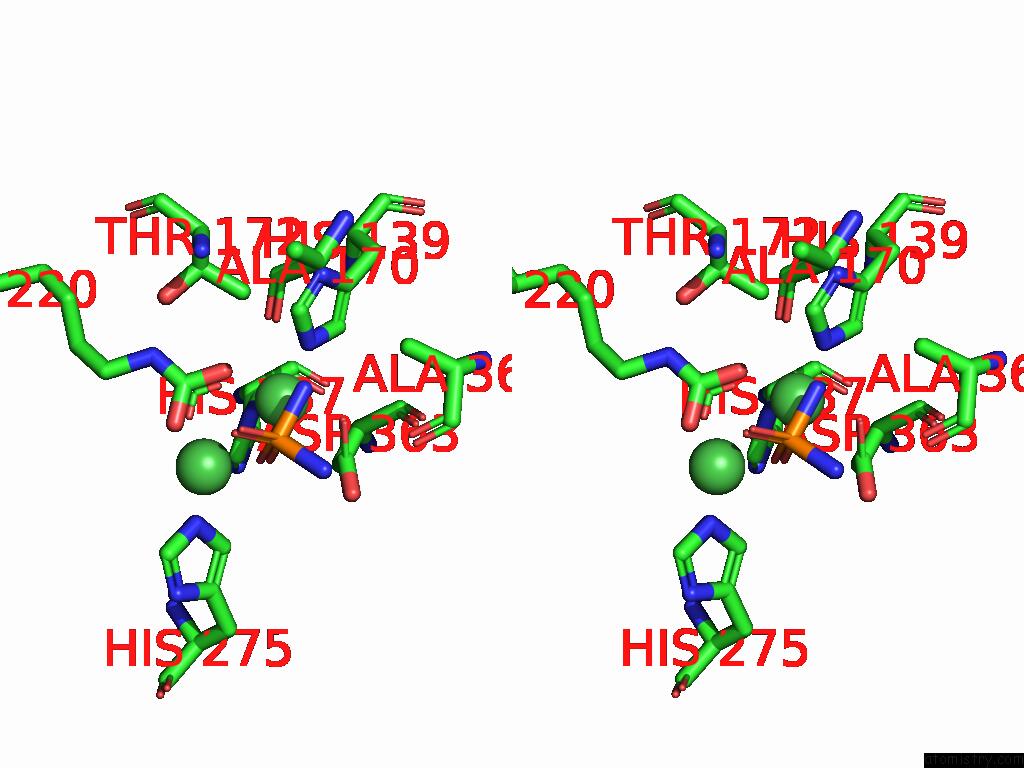
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Cryo-Em Structure of Sporosarcina Pasteurii Urease Inhibited By Nbpto within 5.0Å range:
|
Reference:
L.Mazzei,
G.Tria,
S.Ciurli,
M.Cianci.
Exploring the Conformational Space of the Mobile Flap in Sporosarcina Pasteurii Urease By Cryo-Electron Microscopy. Int.J.Biol.Macromol. V. 283 37904 2024.
ISSN: ISSN 0141-8130
PubMed: 39571870
DOI: 10.1016/J.IJBIOMAC.2024.137904
Page generated: Mon Aug 18 22:37:13 2025
ISSN: ISSN 0141-8130
PubMed: 39571870
DOI: 10.1016/J.IJBIOMAC.2024.137904
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01