Nickel »
PDB 1a5n-1fwb »
1e3d »
Nickel in PDB 1e3d: [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774
Protein crystallography data
The structure of [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774, PDB code: 1e3d
was solved by
P.M.Matias,
C.M.Soares,
L.M.Saraiva,
R.Coelho,
J.Morais,
J.Le Gall,
M.A.Carrondo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 1.80 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.090, 169.980, 72.620, 90.00, 92.80, 90.00 |
| R / Rfree (%) | 16.7 / 22.3 |
Other elements in 1e3d:
The structure of [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774 also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
| Iron | (Fe) | 24 atoms |
Nickel Binding Sites:
The binding sites of Nickel atom in the [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774
(pdb code 1e3d). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774, PDB code: 1e3d:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774, PDB code: 1e3d:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 1e3d
Go back to
Nickel binding site 1 out
of 2 in the [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774
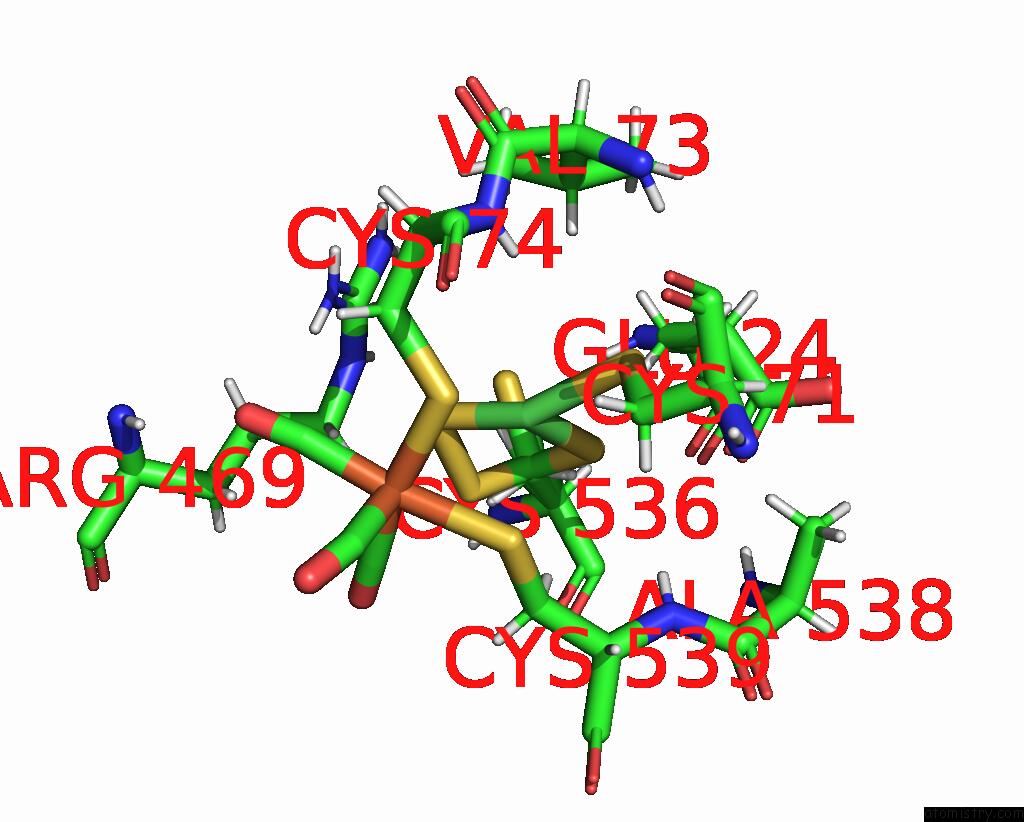
Mono view
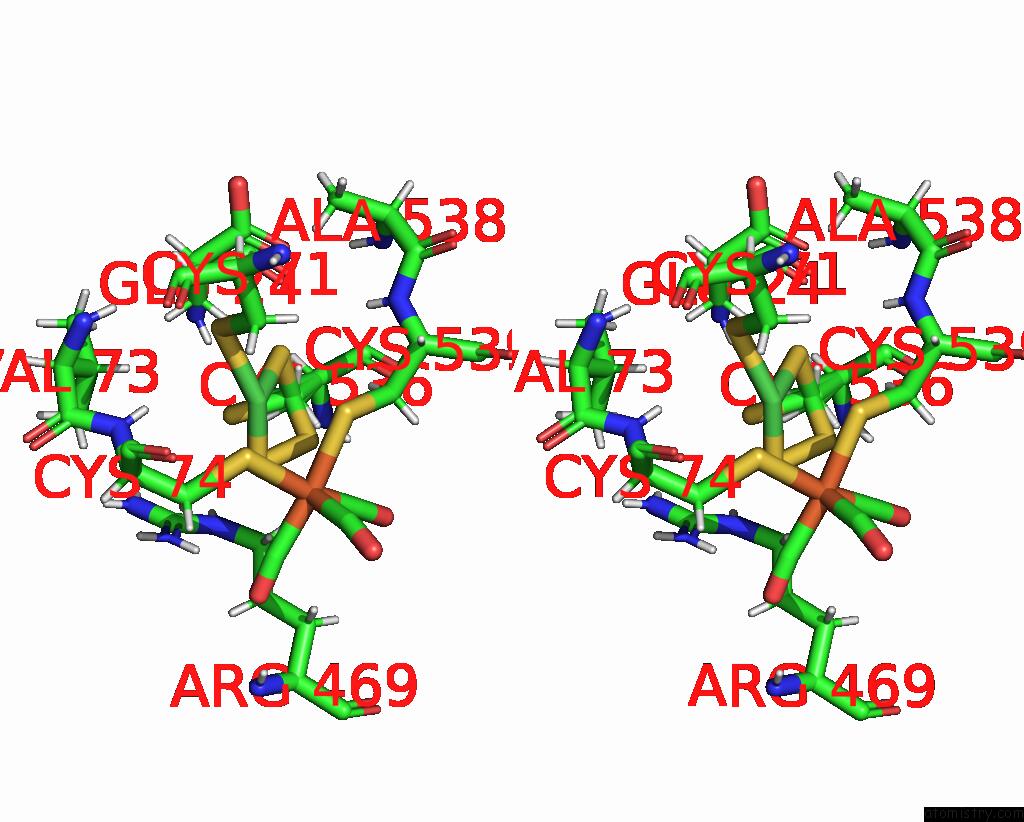
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774 within 5.0Å range:
|
Nickel binding site 2 out of 2 in 1e3d
Go back to
Nickel binding site 2 out
of 2 in the [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774
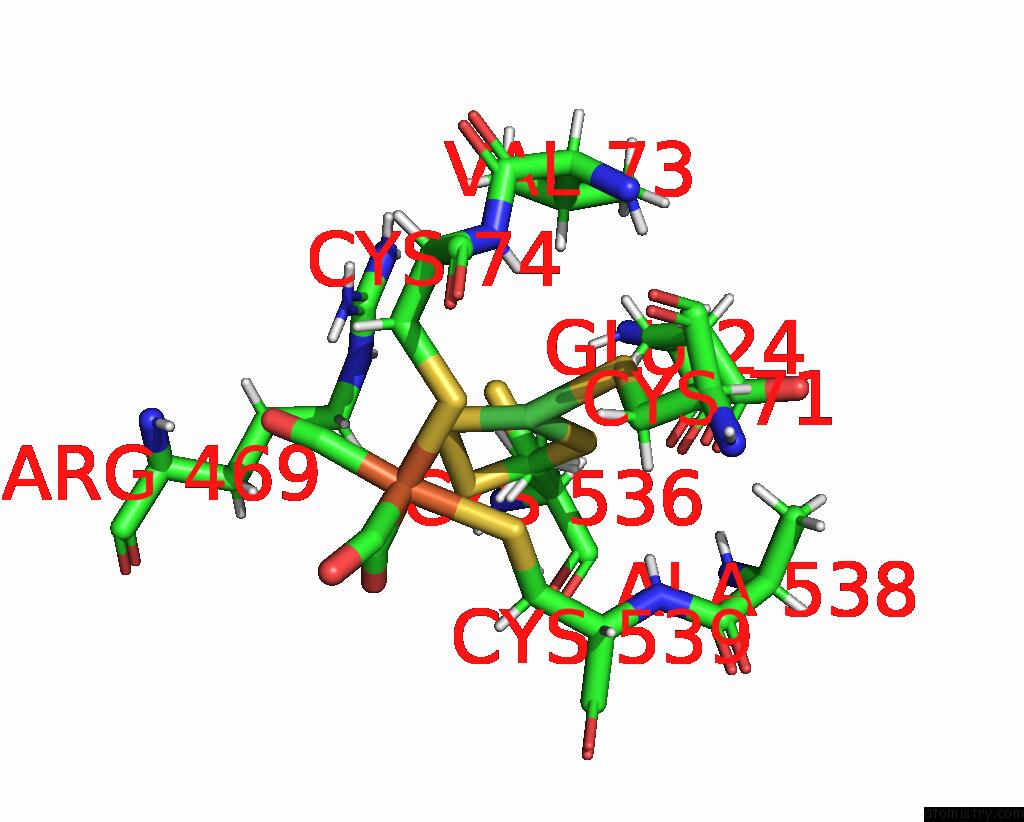
Mono view
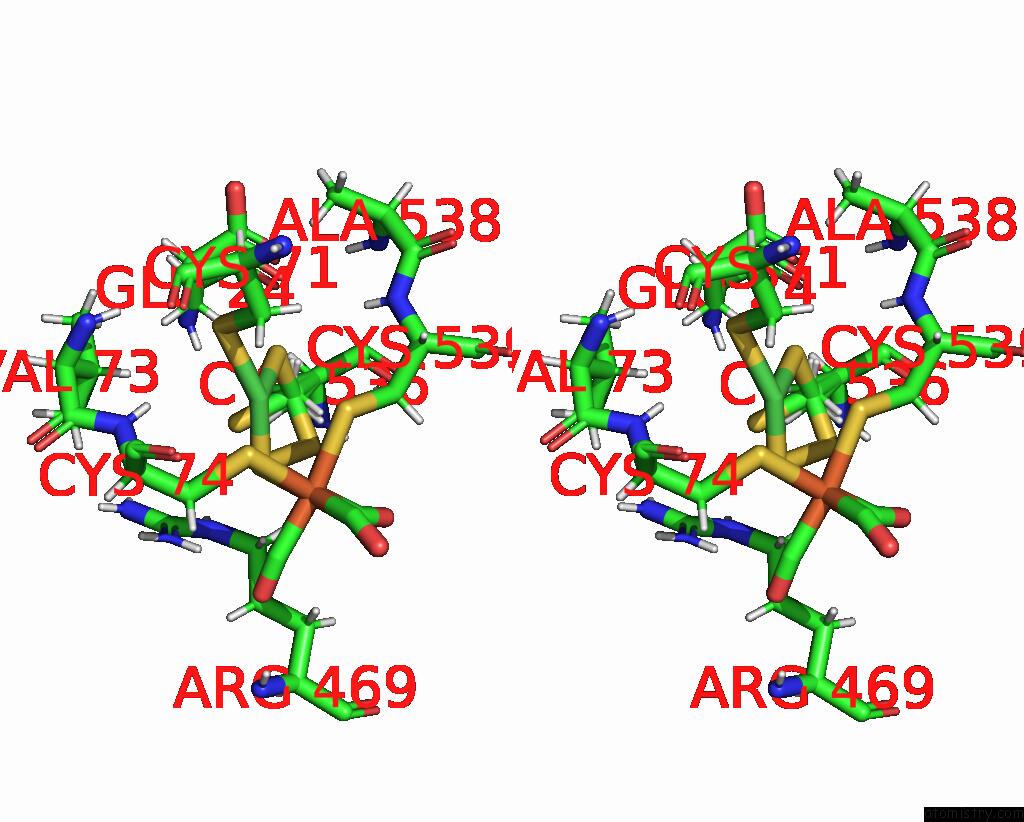
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of [Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774 within 5.0Å range:
|
Reference:
P.M.Matias,
C.M.Soares,
L.M.Saraiva,
R.Coelho,
J.Morais,
J.Le Gall,
M.A.Carrondo.
[Nife] Hydrogenase From Desulfovibrio Desulfuricans Atcc 27774: Gene Sequencing, Three-Dimensional Structure Determination and Refinement at 1.8 A and Modelling Studies of Its Interaction with the Tetrahaem Cytochrome C3. J.Biol.Inorg.Chem. V. 6 63 2001.
ISSN: ISSN 0949-8257
PubMed: 11191224
DOI: 10.1007/S007750000167
Page generated: Mon Aug 18 17:37:19 2025
ISSN: ISSN 0949-8257
PubMed: 11191224
DOI: 10.1007/S007750000167
Last articles
Ni in 5JPTNi in 5JLX
Ni in 5JOV
Ni in 5JLW
Ni in 5JOU
Ni in 5JF0
Ni in 5JJT
Ni in 5JEY
Ni in 5JIG
Ni in 5J7M