Nickel »
PDB 1a5n-1fwb »
1eje »
Nickel in PDB 1eje: Crystal Structure of An Fmn-Binding Protein
Protein crystallography data
The structure of Crystal Structure of An Fmn-Binding Protein, PDB code: 1eje
was solved by
D.Christendat,
V.Saridakis,
A.Bochkarev,
C.Arrowsmith,
A.M.Edwards,
Northeast Structural Genomics Consortium (Nesg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 18.93 / 2.20 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.650, 94.650, 45.400, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.5 / 27.9 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Crystal Structure of An Fmn-Binding Protein
(pdb code 1eje). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the Crystal Structure of An Fmn-Binding Protein, PDB code: 1eje:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the Crystal Structure of An Fmn-Binding Protein, PDB code: 1eje:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 1eje
Go back to
Nickel binding site 1 out
of 2 in the Crystal Structure of An Fmn-Binding Protein
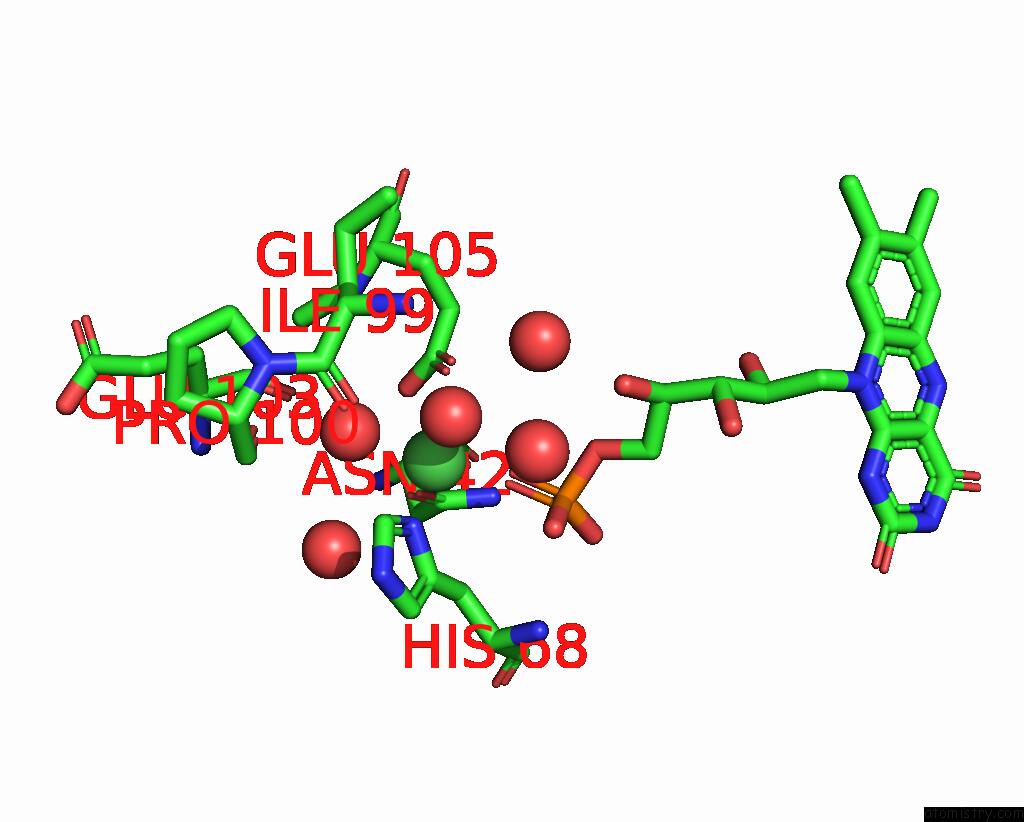
Mono view
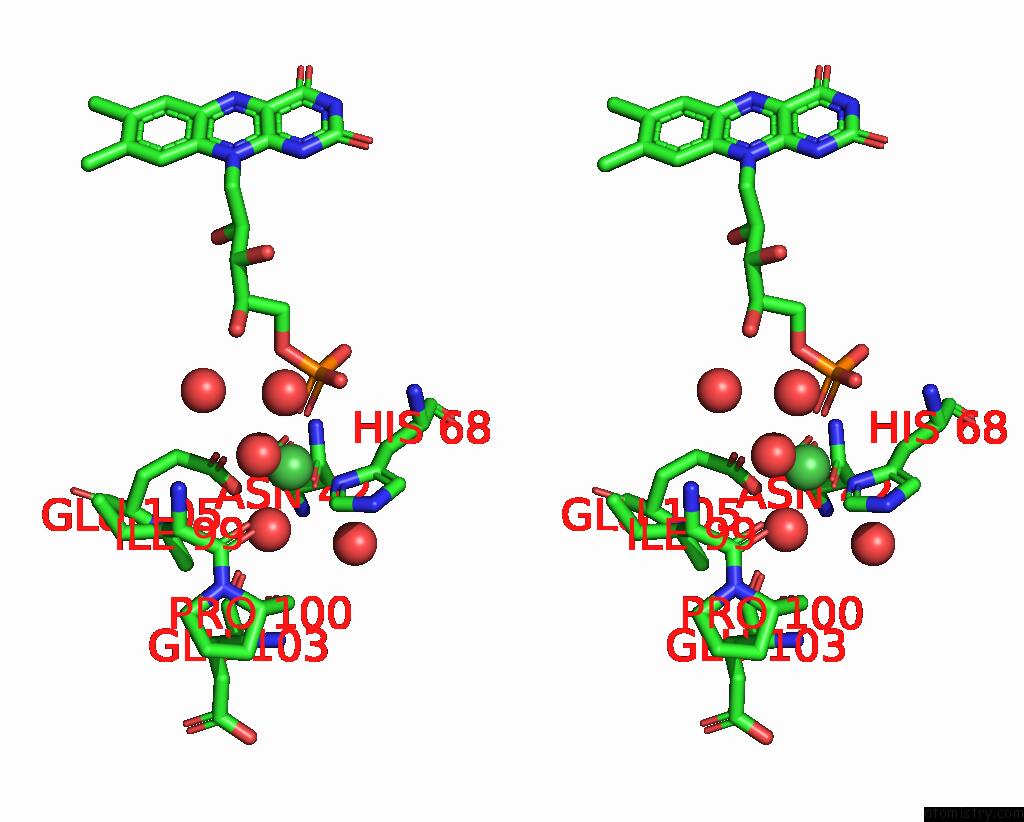
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Crystal Structure of An Fmn-Binding Protein within 5.0Å range:
|
Nickel binding site 2 out of 2 in 1eje
Go back to
Nickel binding site 2 out
of 2 in the Crystal Structure of An Fmn-Binding Protein
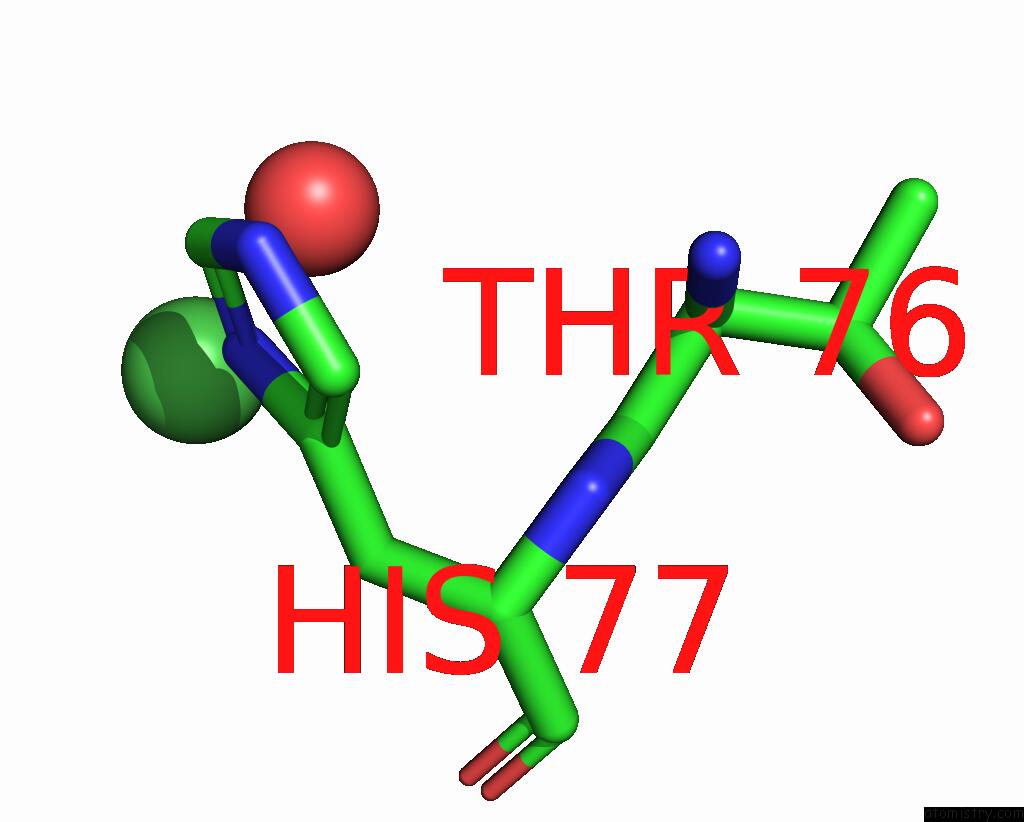
Mono view
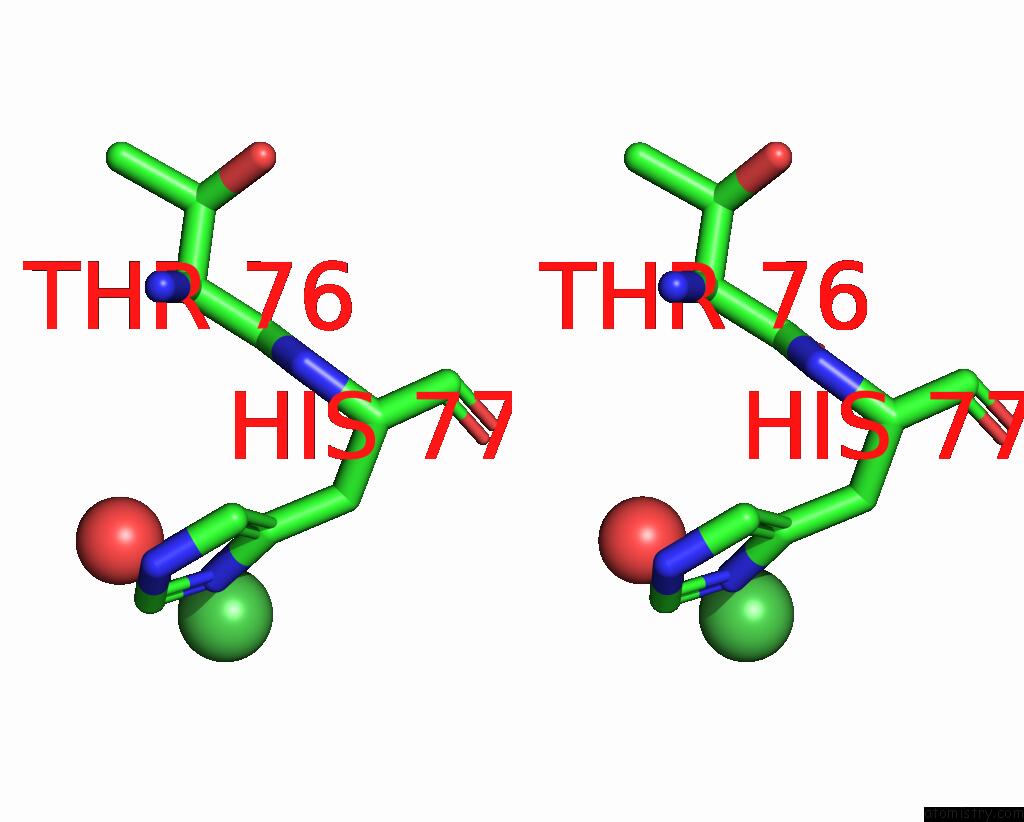
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Crystal Structure of An Fmn-Binding Protein within 5.0Å range:
|
Reference:
D.Christendat,
A.Yee,
A.Dharamsi,
Y.Kluger,
A.Savchenko,
J.R.Cort,
V.Booth,
C.D.Mackereth,
V.Saridakis,
I.Ekiel,
G.Kozlov,
K.L.Maxwell,
N.Wu,
L.P.Mcintosh,
K.Gehring,
M.A.Kennedy,
A.R.Davidson,
E.F.Pai,
M.Gerstein,
A.M.Edwards,
C.H.Arrowsmith.
Structural Proteomics of An Archaeon. Nat.Struct.Biol. V. 7 903 2000.
ISSN: ISSN 1072-8368
PubMed: 11017201
DOI: 10.1038/82823
Page generated: Mon Aug 18 17:38:30 2025
ISSN: ISSN 1072-8368
PubMed: 11017201
DOI: 10.1038/82823
Last articles
Ni in 5NZZNi in 5OA7
Ni in 5OA4
Ni in 5O9W
Ni in 5O7Y
Ni in 5O5D
Ni in 5O59
Ni in 5NZY
Ni in 5NZX
Ni in 5NZW