Nickel »
PDB 1jhj-1q0d »
1oie »
Nickel in PDB 1oie: 5'-Nucleotidase (E. Coli) with An Engineered Disulfide Bridge (S228C, P513C)
Enzymatic activity of 5'-Nucleotidase (E. Coli) with An Engineered Disulfide Bridge (S228C, P513C)
All present enzymatic activity of 5'-Nucleotidase (E. Coli) with An Engineered Disulfide Bridge (S228C, P513C):
3.1.3.5; 3.6.1.45;
3.1.3.5; 3.6.1.45;
Protein crystallography data
The structure of 5'-Nucleotidase (E. Coli) with An Engineered Disulfide Bridge (S228C, P513C), PDB code: 1oie
was solved by
R.Schultz-Heienbrok,
T.Maier,
N.Straeter,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 74.54 / 2.33 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 83.132, 83.132, 180.664, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.8 / 22 |
Nickel Binding Sites:
The binding sites of Nickel atom in the 5'-Nucleotidase (E. Coli) with An Engineered Disulfide Bridge (S228C, P513C)
(pdb code 1oie). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total only one binding site of Nickel was determined in the 5'-Nucleotidase (E. Coli) with An Engineered Disulfide Bridge (S228C, P513C), PDB code: 1oie:
In total only one binding site of Nickel was determined in the 5'-Nucleotidase (E. Coli) with An Engineered Disulfide Bridge (S228C, P513C), PDB code: 1oie:
Nickel binding site 1 out of 1 in 1oie
Go back to
Nickel binding site 1 out
of 1 in the 5'-Nucleotidase (E. Coli) with An Engineered Disulfide Bridge (S228C, P513C)
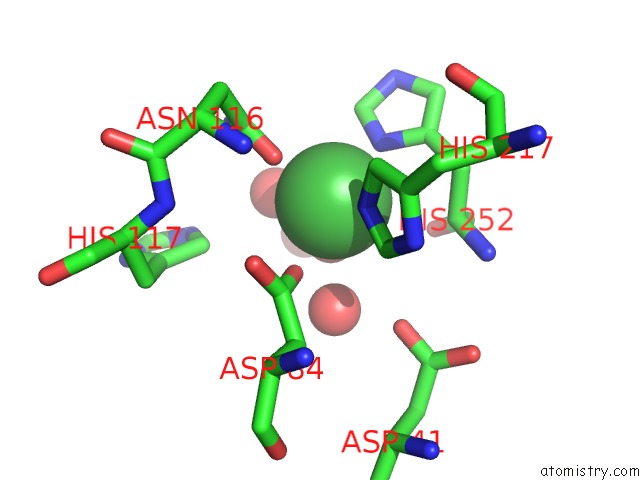
Mono view
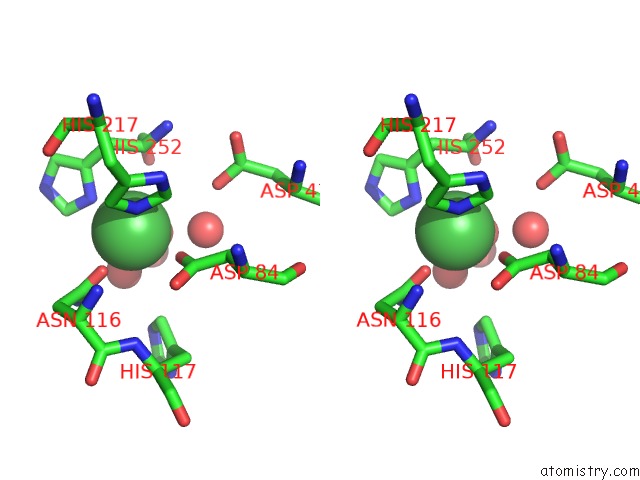
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of 5'-Nucleotidase (E. Coli) with An Engineered Disulfide Bridge (S228C, P513C) within 5.0Å range:
|
Reference:
R.Schultz-Heienbrok,
T.Maier,
N.Straeter.
Trapping A 96 Degree Domain Rotation in Two Distinct Conformations By Engineered Disulfide Bridges Protein Sci. V. 13 1811 2004.
ISSN: ISSN 0961-8368
PubMed: 15215524
DOI: 10.1110/PS.04629604
Page generated: Wed Oct 9 15:04:11 2024
ISSN: ISSN 0961-8368
PubMed: 15215524
DOI: 10.1110/PS.04629604
Last articles
Al in 8UQNAl in 8TSI
Al in 8T0L
Al in 8SHO
Al in 8SHP
Al in 8SHN
Al in 8SHL
Al in 8SHE
Al in 8SHG
Al in 8SHF