Nickel »
PDB 4w6f-4z42 »
4xds »
Nickel in PDB 4xds: Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel
Enzymatic activity of Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel
All present enzymatic activity of Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel:
3.1.5.1;
3.1.5.1;
Protein crystallography data
The structure of Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel, PDB code: 4xds
was solved by
D.Singh,
D.Gawel,
M.Itsko,
J.M.Krahn,
R.E.London,
R.M.Schaaper,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.99 / 3.35 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 189.800, 189.800, 296.800, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.2 / 20.9 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel
(pdb code 4xds). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 6 binding sites of Nickel where determined in the Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel, PDB code: 4xds:
Jump to Nickel binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Nickel where determined in the Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel, PDB code: 4xds:
Jump to Nickel binding site number: 1; 2; 3; 4; 5; 6;
Nickel binding site 1 out of 6 in 4xds
Go back to
Nickel binding site 1 out
of 6 in the Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel
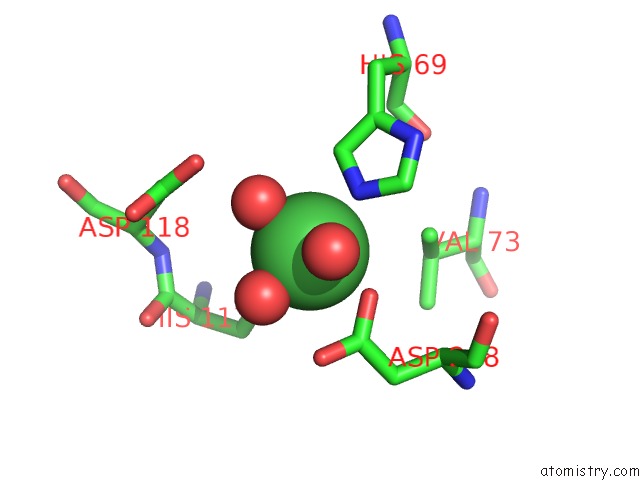
Mono view
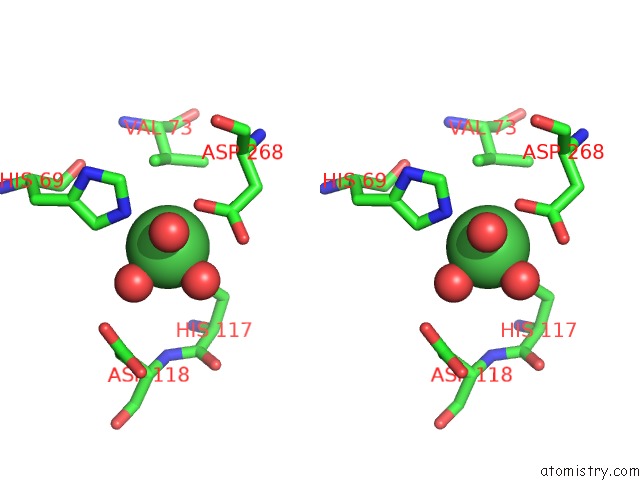
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel within 5.0Å range:
|
Nickel binding site 2 out of 6 in 4xds
Go back to
Nickel binding site 2 out
of 6 in the Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel
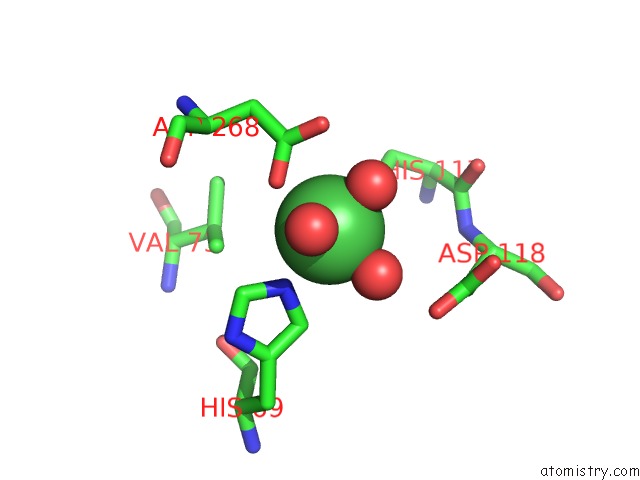
Mono view
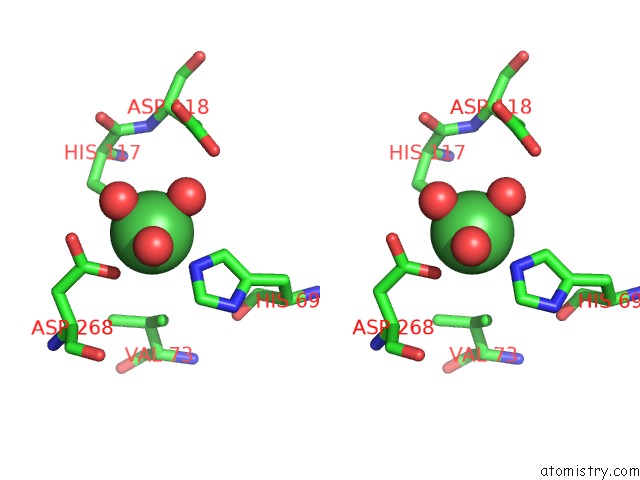
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel within 5.0Å range:
|
Nickel binding site 3 out of 6 in 4xds
Go back to
Nickel binding site 3 out
of 6 in the Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel
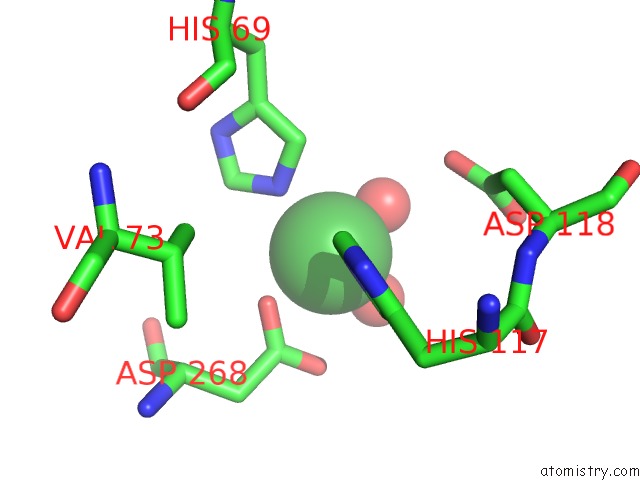
Mono view
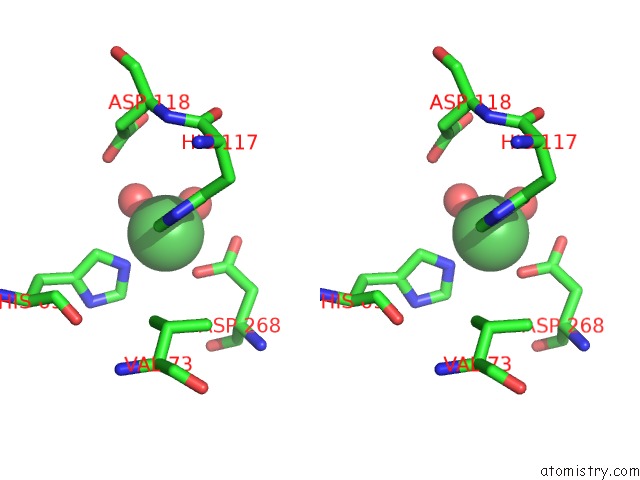
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 3 of Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel within 5.0Å range:
|
Nickel binding site 4 out of 6 in 4xds
Go back to
Nickel binding site 4 out
of 6 in the Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel
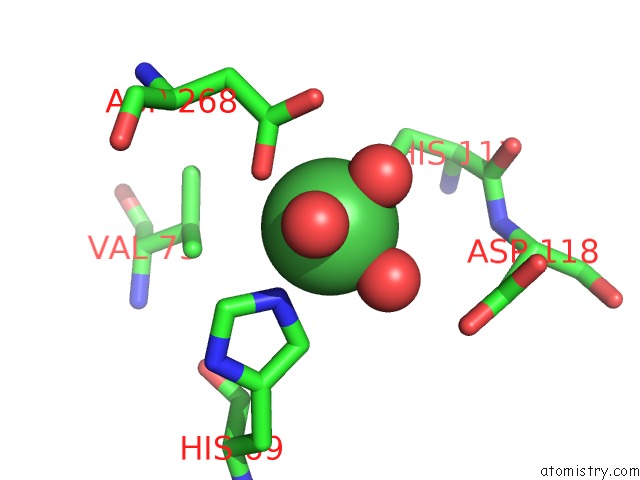
Mono view
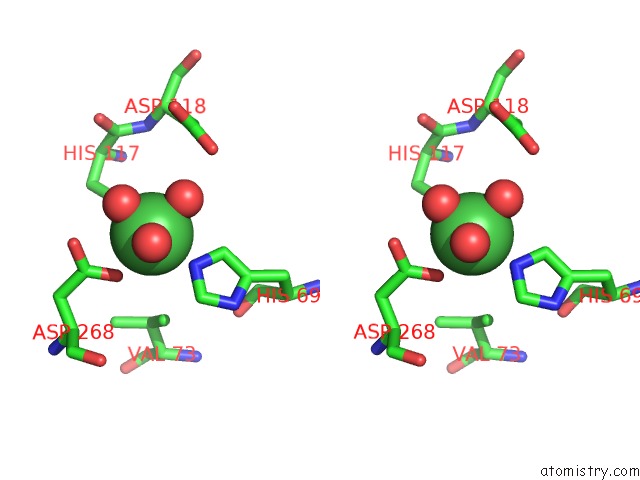
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 4 of Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel within 5.0Å range:
|
Nickel binding site 5 out of 6 in 4xds
Go back to
Nickel binding site 5 out
of 6 in the Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel
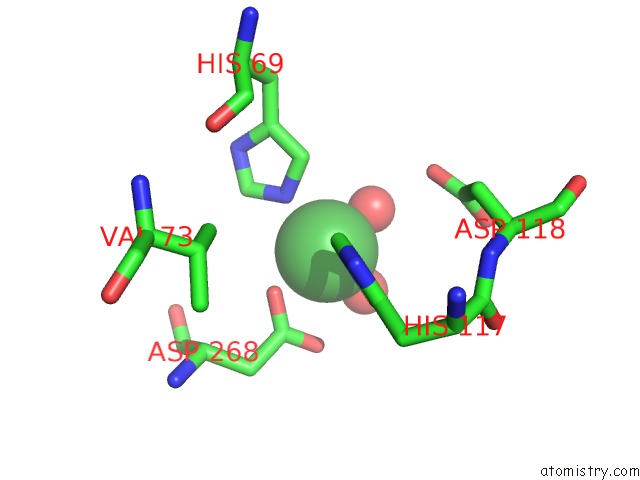
Mono view
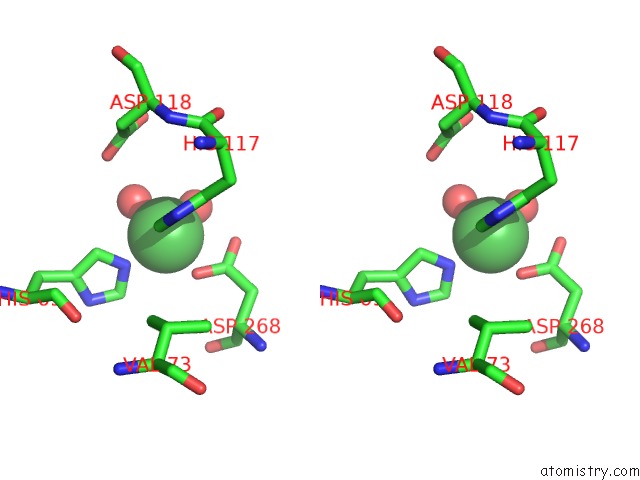
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 5 of Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel within 5.0Å range:
|
Nickel binding site 6 out of 6 in 4xds
Go back to
Nickel binding site 6 out
of 6 in the Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel
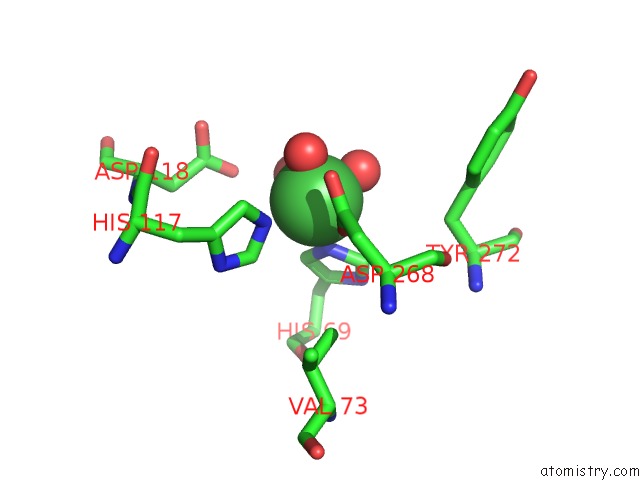
Mono view
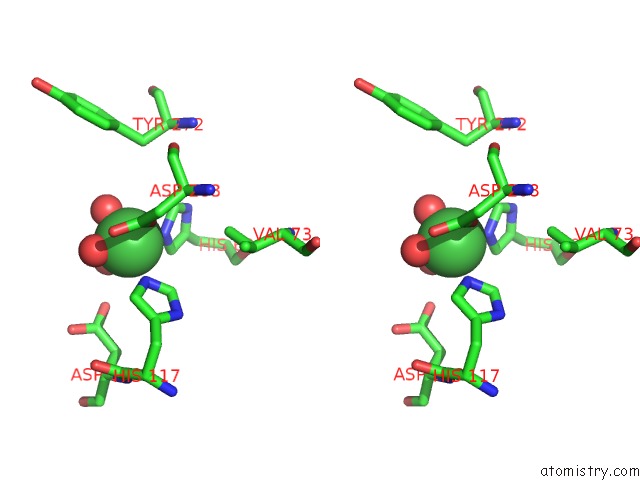
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 6 of Deoxyguanosinetriphosphate Triphosphohydrolase From Escherichia Coli with Nickel within 5.0Å range:
|
Reference:
D.Singh,
D.Gawel,
M.Itsko,
A.Hochkoeppler,
J.M.Krahn,
R.E.London,
R.M.Schaaper.
Structure of Escherichia Coli Dgtp Triphosphohydrolase: A Hexameric Enzyme with Dna Effector Molecules. J.Biol.Chem. 2015.
ISSN: ESSN 1083-351X
PubMed: 25694425
DOI: 10.1074/JBC.M115.636936
Page generated: Mon Aug 18 19:49:00 2025
ISSN: ESSN 1083-351X
PubMed: 25694425
DOI: 10.1074/JBC.M115.636936
Last articles
Ni in 9IYNNi in 9IYM
Ni in 9IYL
Ni in 9GOF
Ni in 9HI3
Ni in 9H1L
Ni in 9GP1
Ni in 9GNR
Ni in 9GV8
Ni in 9G7I