Nickel »
PDB 4z8g-5bnc »
4ziu »
Nickel in PDB 4ziu: Crystal Structure of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning the Residues From Domain MG7 to the C-Terminus.
Protein crystallography data
The structure of Crystal Structure of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning the Residues From Domain MG7 to the C-Terminus., PDB code: 4ziu
was solved by
I.Garcia-Ferrer,
P.Arede,
J.Gomez-Blanco,
D.Luque,
S.Duquerroy,
J.R.Caston,
T.Goulas,
X.F.Gomis-Ruth,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.98 / 2.70 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 86.720, 136.190, 172.800, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19 / 24 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Crystal Structure of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning the Residues From Domain MG7 to the C-Terminus.
(pdb code 4ziu). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 2 binding sites of Nickel where determined in the Crystal Structure of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning the Residues From Domain MG7 to the C-Terminus., PDB code: 4ziu:
Jump to Nickel binding site number: 1; 2;
In total 2 binding sites of Nickel where determined in the Crystal Structure of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning the Residues From Domain MG7 to the C-Terminus., PDB code: 4ziu:
Jump to Nickel binding site number: 1; 2;
Nickel binding site 1 out of 2 in 4ziu
Go back to
Nickel binding site 1 out
of 2 in the Crystal Structure of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning the Residues From Domain MG7 to the C-Terminus.
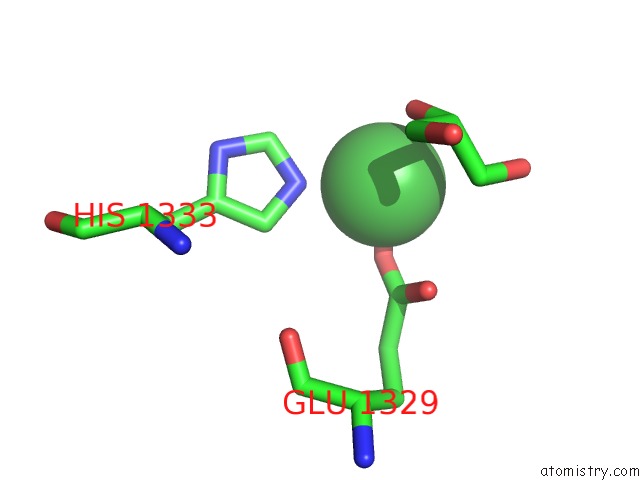
Mono view
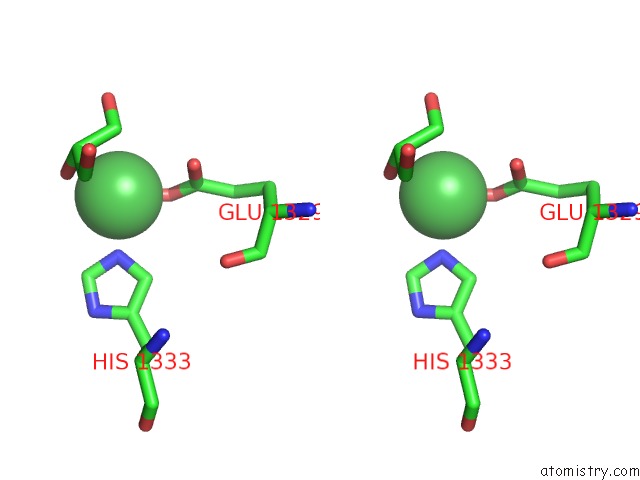
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Crystal Structure of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning the Residues From Domain MG7 to the C-Terminus. within 5.0Å range:
|
Nickel binding site 2 out of 2 in 4ziu
Go back to
Nickel binding site 2 out
of 2 in the Crystal Structure of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning the Residues From Domain MG7 to the C-Terminus.
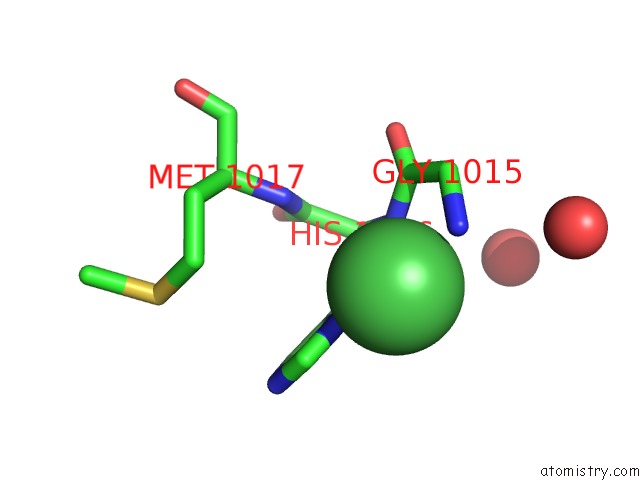
Mono view
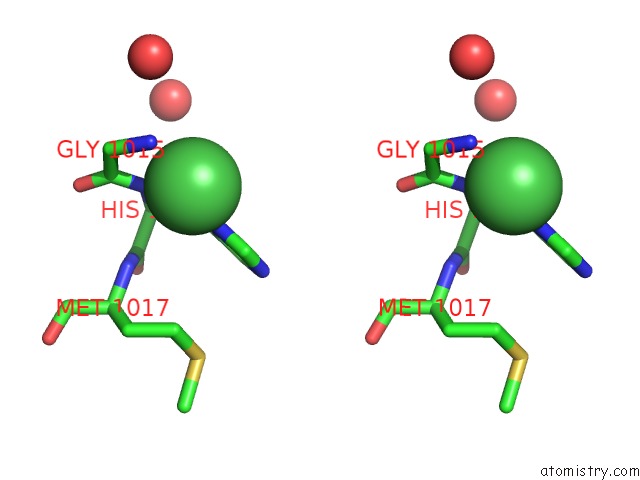
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Crystal Structure of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning the Residues From Domain MG7 to the C-Terminus. within 5.0Å range:
|
Reference:
I.Garcia-Ferrer,
P.Arede,
J.Gomez-Blanco,
D.Luque,
S.Duquerroy,
J.R.Caston,
T.Goulas,
F.X.Gomis-Ruth.
Structural and Functional Insights Into Escherichia Coli Alpha 2-Macroglobulin Endopeptidase Snap-Trap Inhibition. Proc.Natl.Acad.Sci.Usa V. 112 8290 2015.
ISSN: ESSN 1091-6490
PubMed: 26100869
DOI: 10.1073/PNAS.1506538112
Page generated: Mon Aug 18 19:52:36 2025
ISSN: ESSN 1091-6490
PubMed: 26100869
DOI: 10.1073/PNAS.1506538112
Last articles
Ni in 9IYONi in 9IYN
Ni in 9IYM
Ni in 9IYL
Ni in 9GOF
Ni in 9HI3
Ni in 9H1L
Ni in 9GP1
Ni in 9GNR
Ni in 9GV8