Nickel »
PDB 8zak-9gp1 »
9b06 »
Nickel in PDB 9b06: Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+
Protein crystallography data
The structure of Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+, PDB code: 9b06
was solved by
C.J.Presloid,
J.Jiang,
P.C.Beardslee,
H.R.Anderson,
T.M.Swayne,
K.R.Schmitz,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.62 / 1.75 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.244, 52.707, 54.044, 90, 90, 90 |
| R / Rfree (%) | 14.1 / 18.9 |
Other elements in 9b06:
The structure of Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+ also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
Nickel Binding Sites:
The binding sites of Nickel atom in the Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+
(pdb code 9b06). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total 3 binding sites of Nickel where determined in the Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+, PDB code: 9b06:
Jump to Nickel binding site number: 1; 2; 3;
In total 3 binding sites of Nickel where determined in the Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+, PDB code: 9b06:
Jump to Nickel binding site number: 1; 2; 3;
Nickel binding site 1 out of 3 in 9b06
Go back to
Nickel binding site 1 out
of 3 in the Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+
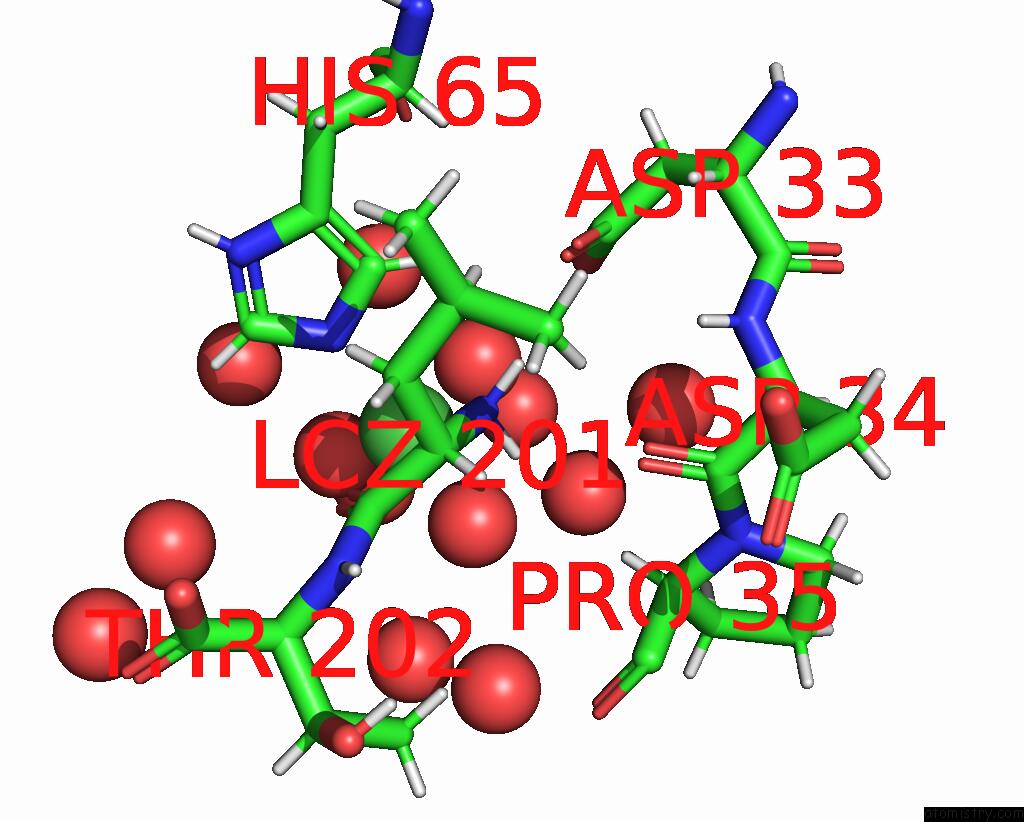
Mono view
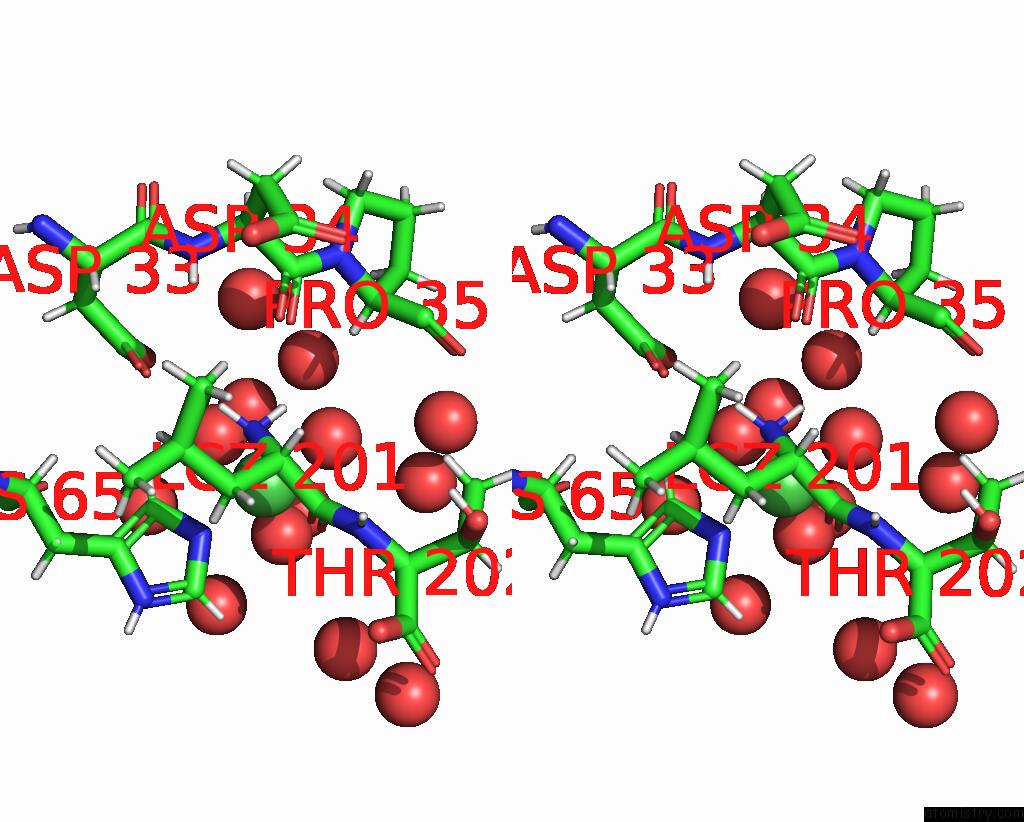
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+ within 5.0Å range:
|
Nickel binding site 2 out of 3 in 9b06
Go back to
Nickel binding site 2 out
of 3 in the Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+
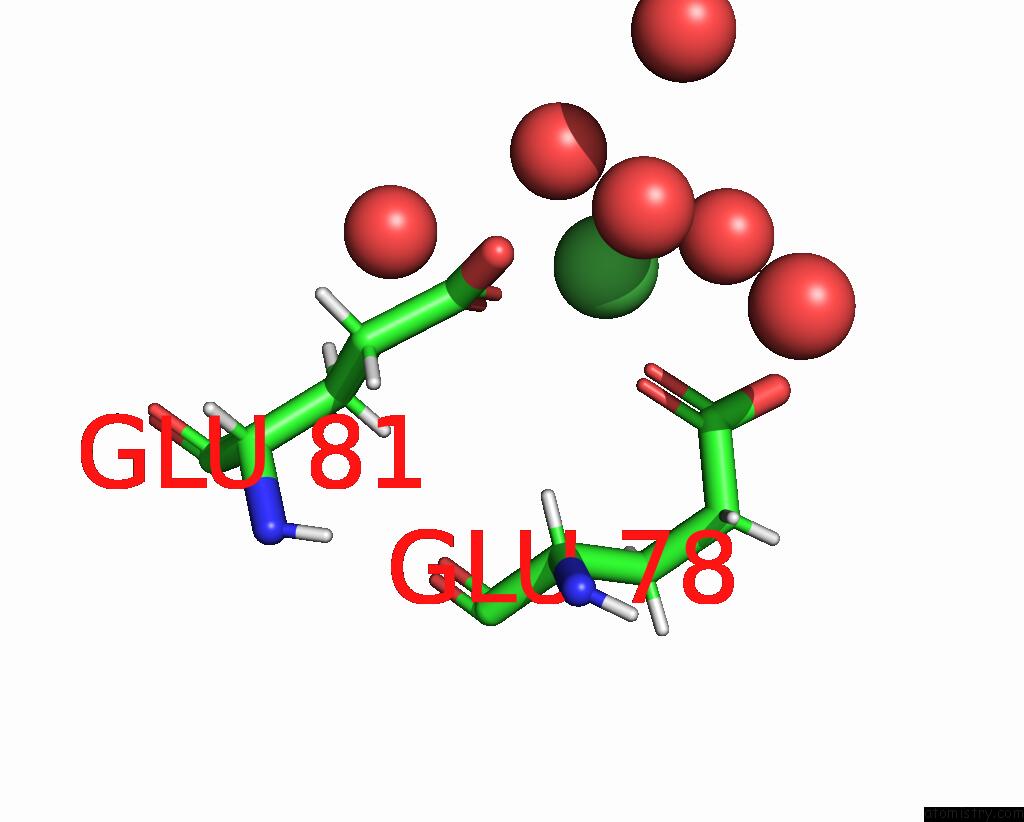
Mono view
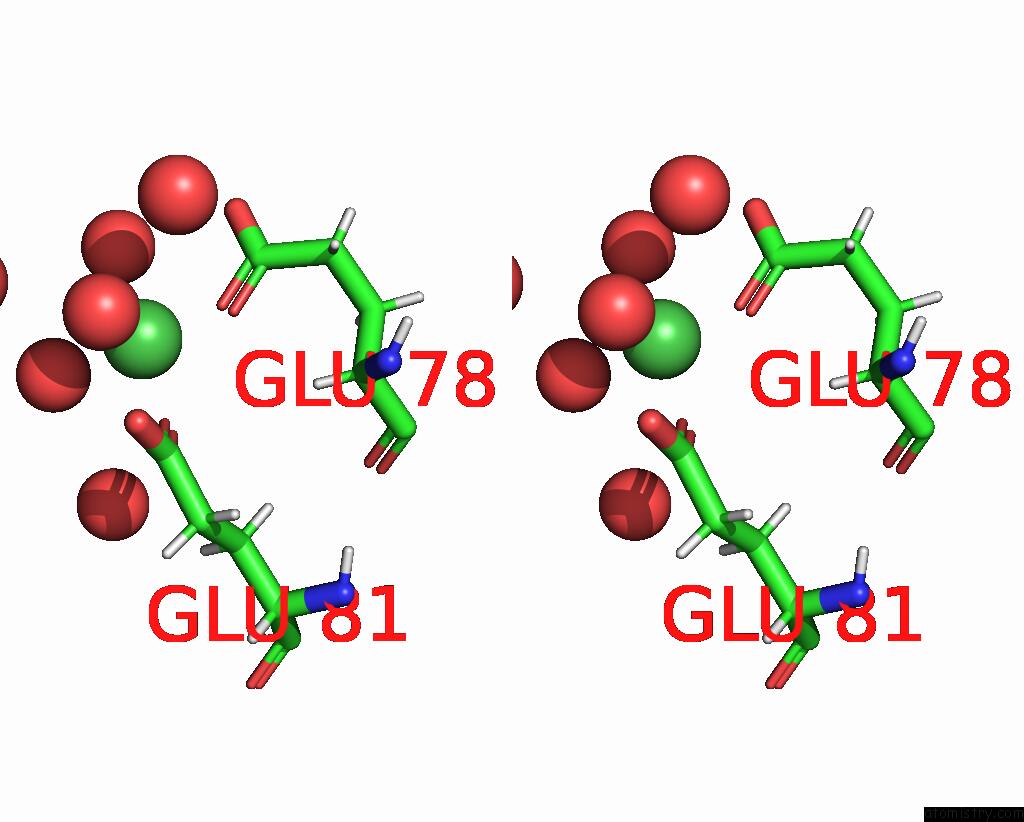
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 2 of Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+ within 5.0Å range:
|
Nickel binding site 3 out of 3 in 9b06
Go back to
Nickel binding site 3 out
of 3 in the Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+
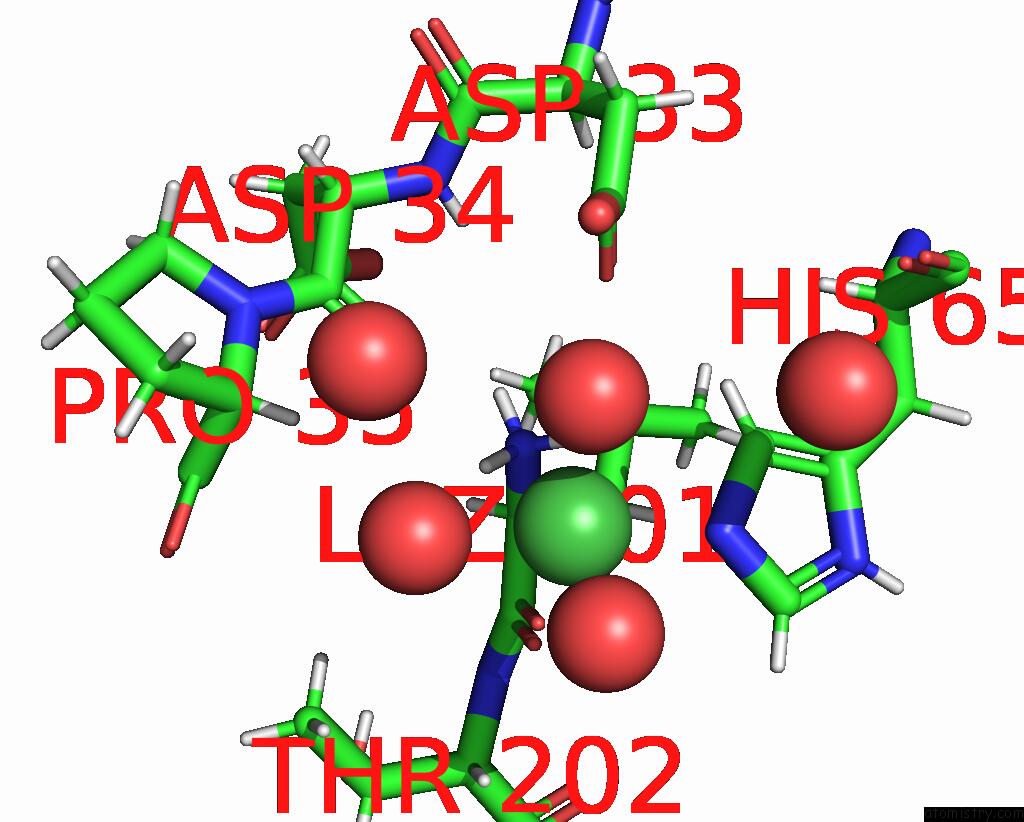
Mono view
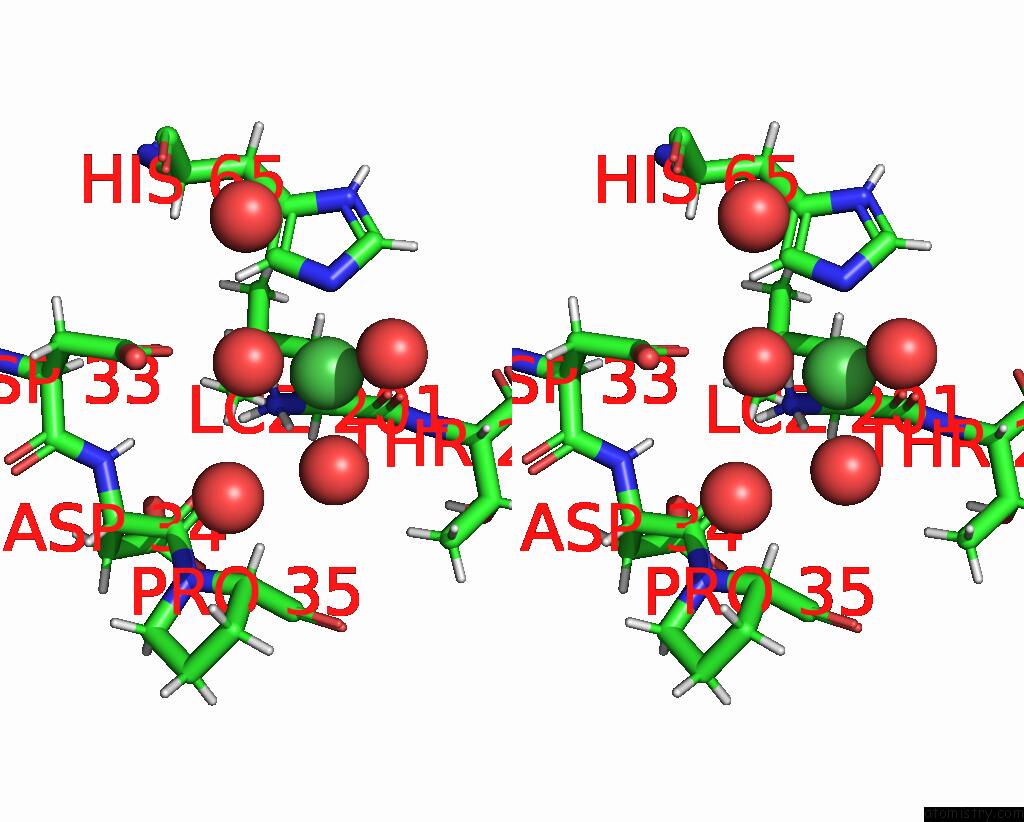
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 3 of Mycolicibacterium Smegmatis Clps with Leuthr Dipeptide and MG2+ within 5.0Å range:
|
Reference:
C.J.Presloid,
J.Jiang,
P.Kandel,
H.R.Anderson,
P.C.Beardslee,
T.M.Swayne,
K.R.Schmitz.
Clps Directs Degradation of N-Degron Substrates with Primary Destabilizing Residues in Mycolicibacterium Smegmatis. Mol.Microbiol. 2024.
ISSN: ESSN 1365-2958
PubMed: 39626090
DOI: 10.1111/MMI.15334
Page generated: Sun Dec 15 11:47:32 2024
ISSN: ESSN 1365-2958
PubMed: 39626090
DOI: 10.1111/MMI.15334
Last articles
I in 9AXJHg in 9D67
Hg in 9D66
Hg in 9D69
Hg in 9D6A
Fe in 9J2F
Fe in 9JDC
Fe in 9JDB
Fe in 9KU6
Fe in 9F47