Nickel »
PDB 5e6m-5i14 »
5ff5 »
Nickel in PDB 5ff5: Crystal Structure of Semet Paaa
Protein crystallography data
The structure of Crystal Structure of Semet Paaa, PDB code: 5ff5
was solved by
K.B.Biernat,
M.R.Redinbo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.48 / 2.93 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.962, 170.409, 64.323, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21 / 28.9 |
Nickel Binding Sites:
The binding sites of Nickel atom in the Crystal Structure of Semet Paaa
(pdb code 5ff5). This binding sites where shown within
5.0 Angstroms radius around Nickel atom.
In total only one binding site of Nickel was determined in the Crystal Structure of Semet Paaa, PDB code: 5ff5:
In total only one binding site of Nickel was determined in the Crystal Structure of Semet Paaa, PDB code: 5ff5:
Nickel binding site 1 out of 1 in 5ff5
Go back to
Nickel binding site 1 out
of 1 in the Crystal Structure of Semet Paaa
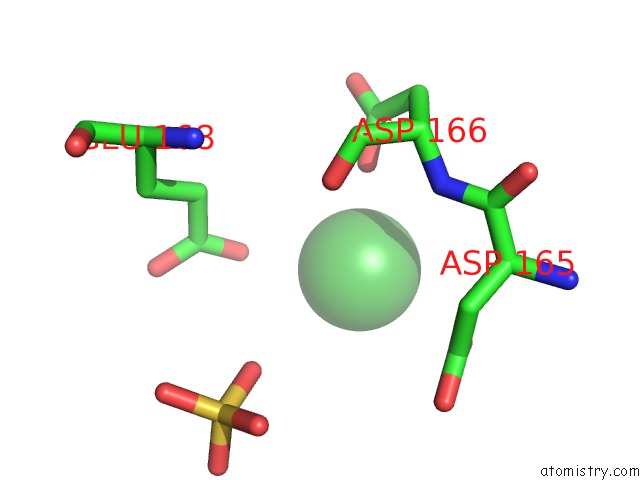
Mono view
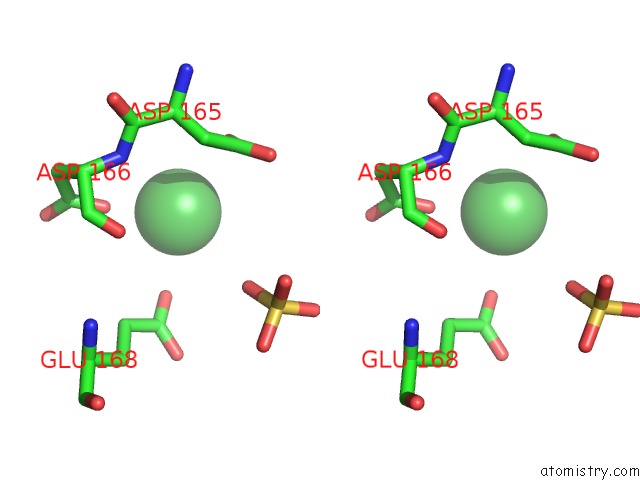
Stereo pair view

Mono view

Stereo pair view
A full contact list of Nickel with other atoms in the Ni binding
site number 1 of Crystal Structure of Semet Paaa within 5.0Å range:
|
Reference:
S.V.Ghodge,
K.A.Biernat,
S.J.Bassett,
M.R.Redinbo,
A.A.Bowers.
Post-Translational Claisen Condensation and Decarboxylation En Route to the Bicyclic Core of Pantocin A. J.Am.Chem.Soc. V. 138 5487 2016.
ISSN: ESSN 1520-5126
PubMed: 27088303
DOI: 10.1021/JACS.5B13529
Page generated: Mon Aug 18 20:01:41 2025
ISSN: ESSN 1520-5126
PubMed: 27088303
DOI: 10.1021/JACS.5B13529
Last articles
Ni in 9KHONi in 9QM5
Ni in 9KHQ
Ni in 9OFX
Ni in 9NF0
Ni in 9NEZ
Ni in 9MYP
Ni in 9GYB
Ni in 9IYU
Ni in 9IYT